Clinical study of genomic drivers in pancreatic ductal adenocarcinoma
- PMID: 28720843
- PMCID: PMC5558689
- DOI: 10.1038/bjc.2017.209
Clinical study of genomic drivers in pancreatic ductal adenocarcinoma
Abstract
Background: Pancreatic ductal adenocarcinoma (PDA) is a lethal cancer with complex genomes and dense fibrotic stroma. This study was designed to identify clinically relevant somatic aberrations in pancreatic cancer genomes of patients with primary and metastatic disease enrolled and treated in two clinical trials.
Methods: Tumour nuclei were flow sorted prior to whole genome copy number variant (CNV) analysis. Targeted or whole exome sequencing was performed on most samples. We profiled biopsies from 68 patients enrolled in two Stand Up to Cancer (SU2C)-sponsored clinical trials. These included 38 resected chemoradiation naïve tumours (SU2C 20206-003) and metastases from 30 patients who progressed on prior therapies (SU2C 20206-001). Patient outcomes including progression-free survival (PFS) and overall survival (OS) were observed.
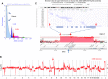
Results: We defined: (a) CDKN2A homozygous deletions that included the adjacent MTAP gene, only its' 3' region, or excluded MTAP; (b) SMAD4 homozygous deletions that included ME2; (c) a pancreas-specific MYC super-enhancer region; (d) DNA repair-deficient genomes; and (e) copy number aberrations present in PDA patients with long-term (⩾ 40 months) and short-term (⩽ 12 months) survival after surgical resection.
Conclusions: We provide a clinically relevant framework for genomic drivers of PDA and for advancing novel treatments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grutzmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM Australian Pancreatic Cancer Genome IMunzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531(7592): 47–52. - PubMed
-
- Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, Dueck AC, McCullough AE, Reddy SK, Ramanathan RK, Northfelt DW, Pockaj BA (2015) Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 6(28): 26483–26493. - PMC - PubMed
-
- Biankin AV, Maitra A (2015) Subtyping pancreatic cancer. Cancer Cell 28(4): 411–413. - PubMed
-
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, Perez-Atayde A, Wong KK, Yuan GC, Gray NS, Young RA, George RE (2014) CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159(5): 1126–1139. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

