A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity
- PMID: 28721243
- PMCID: PMC5515256
- DOI: 10.1038/npjbiofilms.2016.4
A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity
Abstract
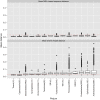
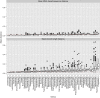
The standard pipeline for 16S amplicon analysis starts by clustering sequences within a percent sequence similarity threshold (typically 97%) into 'Operational Taxonomic Units' (OTUs). From each OTU, a single sequence is selected as a representative. This representative sequence is annotated, and that annotation is applied to all remaining sequences within that OTU. This perspective paper will discuss the known shortcomings of this standard approach using results obtained from the Human Microbiome Project. In particular, we will show that the traditional approach of using pairwise sequence alignments to compute sequence similarity can result in poorly clustered OTUs. As OTUs are typically annotated based upon a single representative sequence, poorly clustered OTUs can have significant impact on downstream analyses. These results suggest that we need to move beyond simple clustering techniques for 16S analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

