Genome analysis of E. coli isolated from Crohn's disease patients
- PMID: 28724357
- PMCID: PMC5517970
- DOI: 10.1186/s12864-017-3917-x
Genome analysis of E. coli isolated from Crohn's disease patients
Abstract
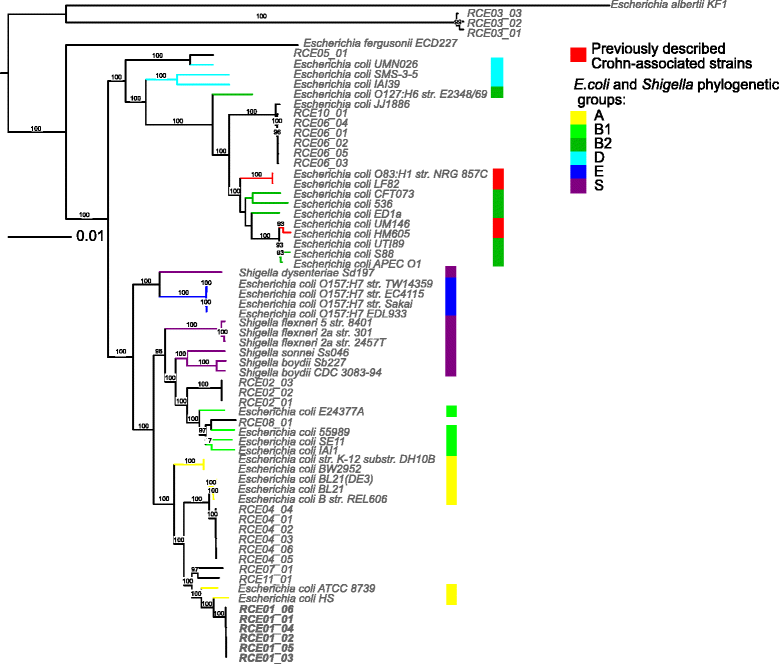
Background: Escherichia coli (E. coli) has been increasingly implicated in the pathogenesis of Crohn's disease (CD). The phylogeny of E. coli isolated from Crohn's disease patients (CDEC) was controversial, and while genotyping results suggested heterogeneity, the sequenced strains of E. coli from CD patients were closely related.
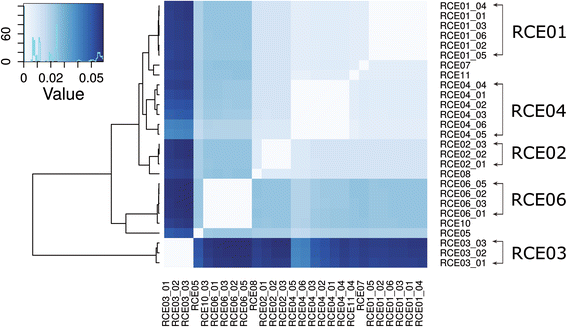
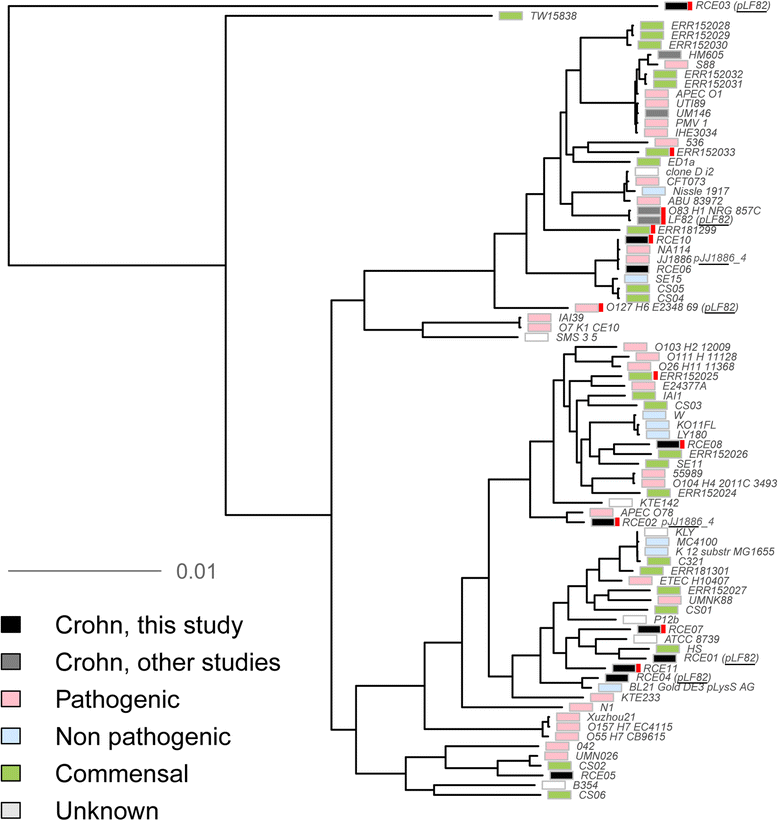
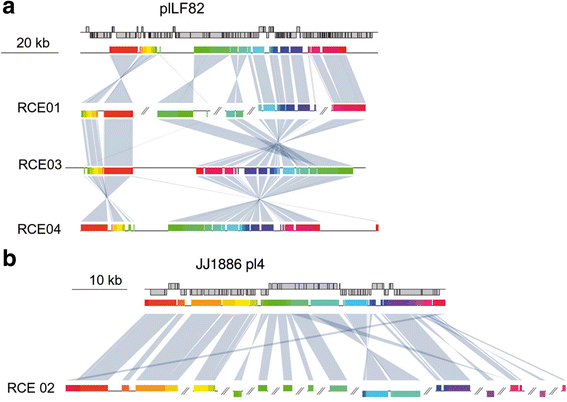
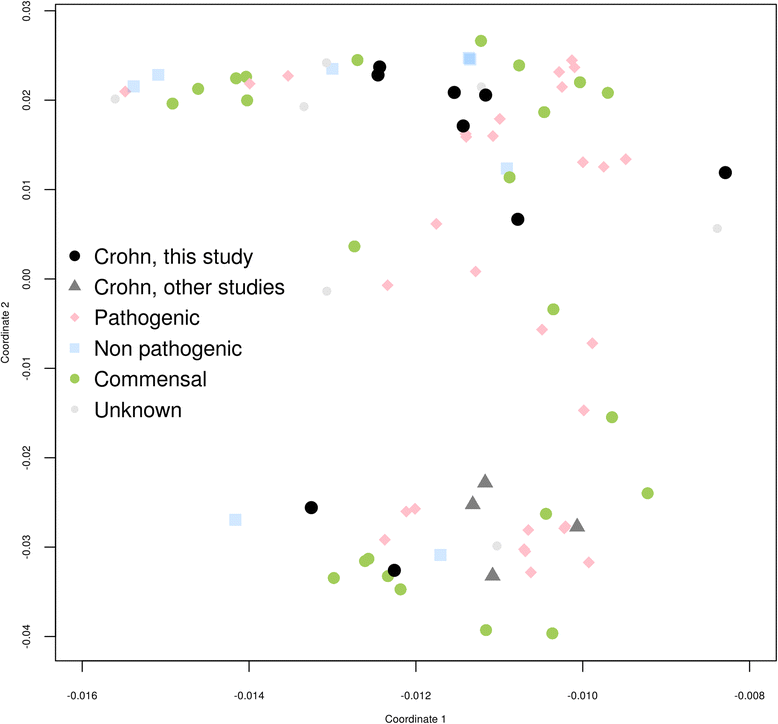
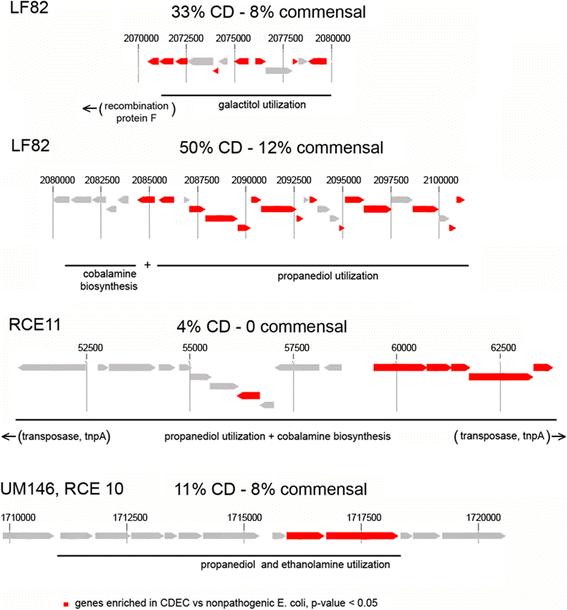
Results: We performed the shotgun genome sequencing of 28 E. coli isolates from ten CD patients and compared genomes from these isolates with already published genomes of CD strains and other pathogenic and non-pathogenic strains. CDEC was shown to belong to A, B1, B2 and D phylogenetic groups. The plasmid and several operons from the reference CD-associated E. coli strain LF82 were demonstrated to be more often present in CDEC genomes belonging to different phylogenetic groups than in genomes of commensal strains. The operons include carbon-source induced invasion GimA island, prophage I, iron uptake operons I and II, capsular assembly pathogenetic island IV and propanediol and galactitol utilization operons.
Conclusions: Our findings suggest that CDEC are phylogenetically diverse. However, some strains isolated from independent sources possess highly similar chromosome or plasmids. Though no CD-specific genes or functional domains were present in all CD-associated strains, some genes and operons are more often found in the genomes of CDEC than in commensal E. coli. They are principally linked to gut colonization and utilization of propanediol and other sugar alcohols.
Keywords: Crohn’s disease; E. coli; Genome; Propanediol.
Conflict of interest statement
Ethics approval and consent to participate
Patient Anonymity and Informed Consent. The study was approved by ethical committees of Central Scientific Institute of Gastroenterology and State Scientific Center of Coloproctology. All patients gave written informed consent for sample collection and personal data processing. All samples were anonymized.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

