ALX receptor ligands define a biochemical endotype for severe asthma
- PMID: 28724795
- PMCID: PMC5518567
- DOI: 10.1172/jci.insight.93534
ALX receptor ligands define a biochemical endotype for severe asthma
Erratum in
-
ALX receptor ligands define a biochemical endotype for severe asthma.JCI Insight. 2018 Mar 22;3(6):e120932. doi: 10.1172/jci.insight.120932. JCI Insight. 2018. PMID: 29563345 Free PMC article. No abstract available.
Abstract
Background: In health, inflammation resolution is an active process governed by specialized proresolving mediators and receptors. ALX/FPR2 receptors (ALX) are targeted by both proresolving and proinflammatory ligands for opposing signaling events, suggesting pivotal roles for ALX in the fate of inflammatory responses. Here, we determined if ALX expression and ligands were linked to severe asthma (SA).
Methods: ALX expression and levels of proresolving ligands (lipoxin A4 [LXA4], 15-epi-LXA4, and annexin A1 [ANXA1]), and a proinflammatory ligand (serum amyloid A [SAA]) were measured in bronchoscopy samples collected in Severe Asthma Research Program-3 (SA [n = 69], non-SA [NSA, n = 51] or healthy donors [HDs, n = 47]).
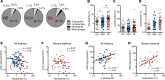
Results: Bronchoalveolar lavage (BAL) fluid LXA4 and 15-epi-LXA4 were decreased and SAA was increased in SA relative to NSA. BAL macrophage ALX expression was increased in SA. Subjects with LXA4loSAAhi levels had increased BAL neutrophils, more asthma symptoms, lower lung function, increased relative risk for asthma exacerbation, sinusitis, and gastroesophageal reflux disease, and were assigned more frequently to SA clinical clusters. SAA and aliquots of LXA4loSAAhi BAL fluid induced IL-8 production by lung epithelial cells expressing ALX receptors, which was inhibited by coincubation with 15-epi-LXA4.
Conclusions: Together, these findings have established an association between select ALX receptor ligands and asthma severity that define a potentially new biochemical endotype for asthma and support a pivotal functional role for ALX signaling in the fate of lung inflammation.
Trial registration: Severe Asthma Research Program-3 (SARP-3; ClinicalTrials.gov NCT01606826)FUNDING Sources. National Heart, Lung and Blood Institute, the NIH, and the German Society of Pediatric Pneumology.
Keywords: Inflammation; Pulmonology.
Conflict of interest statement
Figures





References
-
- Most Recent Asthma Data. Center for Disease Control and Prevention. https://www.cdc.gov/asthma/most_recent_data.htm Updated February 27, 2017. Accessed June 6, 2017.
Associated data
Grants and funding
- U10 HL109257/HL/NHLBI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- K23 HL138303/HL/NHLBI NIH HHS/United States
- U10 HL109250/HL/NHLBI NIH HHS/United States
- R01 HL122531/HL/NHLBI NIH HHS/United States
- U10 HL109164/HL/NHLBI NIH HHS/United States
- U10 HL109172/HL/NHLBI NIH HHS/United States
- U10 HL064313/HL/NHLBI NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- K12 HD047349/HD/NICHD NIH HHS/United States
- U10 HL109086/HL/NHLBI NIH HHS/United States
- U10 HL109168/HL/NHLBI NIH HHS/United States
- UL1 TR001102/TR/NCATS NIH HHS/United States
- U10 HL109152/HL/NHLBI NIH HHS/United States
- U10 HL109146/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

