Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein-Protein Interaction Analysis, and Protein Function Exploration
- PMID: 28725224
- PMCID: PMC5495861
- DOI: 10.3389/fimmu.2017.00771
Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein-Protein Interaction Analysis, and Protein Function Exploration
Abstract
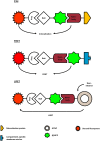
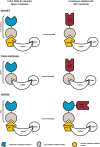
Over the last two decades, nanobodies or single-domain antibodies have found their way in research, diagnostics, and therapy. These antigen-binding fragments, derived from Camelid heavy chain only antibodies, possess remarkable characteristics that favor their use over conventional antibodies or fragments thereof, in selected areas of research. In this review, we assess the current status of nanobodies as research tools in diverse aspects of fundamental research. We discuss the use of nanobodies as detection reagents in fluorescence microscopy and focus on recent advances in super-resolution microscopy. Second, application of nanobody technology in investigating protein-protein interactions is reviewed, with emphasis on possible uses in mass spectrometry. Finally, we discuss the potential value of nanobodies in studying protein function, and we focus on their recently reported application in targeted protein degradation. Throughout the review, we highlight state-of-the-art engineering strategies that could expand nanobody versatility and we suggest future applications of the technology in the selected areas of fundamental research.
Keywords: VHH; engineering; fundamental research; nanobody; protein–protein interactions; single-domain antibody; super-resolution microscopy; targeted protein degradation.
Figures
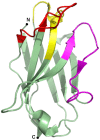

References
-
- Muyldermans S. Nanobodies: natural single-domain antibodies. In: Kornberg RD, editor. Annual Review of Biochemistry. (Vol. 82), Palo Alto: Annual Reviews; (2013). p. 775–97. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

