Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases
- PMID: 28726643
- PMCID: PMC5612095
- DOI: 10.1074/jbc.M117.790865
Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases
Erratum in
-
Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.J Biol Chem. 2017 Dec 22;292(51):21218. doi: 10.1074/jbc.AAC117.001147. J Biol Chem. 2017. PMID: 29273585 Free PMC article. No abstract available.
Abstract
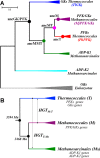
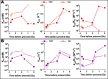
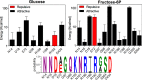
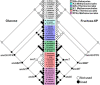
One central goal in molecular evolution is to pinpoint the mechanisms and evolutionary forces that cause an enzyme to change its substrate specificity; however, these processes remain largely unexplored. Using the glycolytic ADP-dependent kinases of archaea, including the orders Thermococcales, Methanosarcinales, and Methanococcales, as a model and employing an approach involving paleoenzymology, evolutionary statistics, and protein structural analysis, we could track changes in substrate specificity during ADP-dependent kinase evolution along with the structural determinants of these changes. To do so, we studied five key resurrected ancestral enzymes as well as their extant counterparts. We found that a major shift in function from a bifunctional ancestor that could phosphorylate either glucose or fructose 6-phosphate (fructose-6-P) as a substrate to a fructose 6-P-specific enzyme was started by a single amino acid substitution resulting in negative selection with a ground-state mode against glucose and a subsequent 1,600-fold change in specificity of the ancestral protein. This change rendered the residual phosphorylation of glucose a promiscuous and physiologically irrelevant activity, highlighting how promiscuity may be an evolutionary vestige of ancestral enzyme activities, which have been eliminated over time. We also could reconstruct the evolutionary history of substrate utilization by using an evolutionary model of discrete binary characters, indicating that substrate uses can be discretely lost or acquired during enzyme evolution. These findings exemplify how negative selection and subtle enzyme changes can lead to major evolutionary shifts in function, which can subsequently generate important adaptive advantages, for example, in improving glycolytic efficiency in Thermococcales.
Keywords: ADP-dependent kinase; ancestral enzymes; archaea; crystal structure; glucokinase; phosphofructokinase; protein evolution; substrate specificity.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Conant G. C., and Wolfe K. H. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950 - PubMed
-
- Todd A. E., Orengo C. A., and Thornton J. M. (2001) Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307, 1113–1143 - PubMed
-
- Khersonsky O., and Tawfik D. S. (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

