The whole-genome landscape of medulloblastoma subtypes
- PMID: 28726821
- PMCID: PMC5905700
- DOI: 10.1038/nature22973
The whole-genome landscape of medulloblastoma subtypes
Abstract
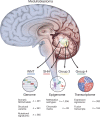
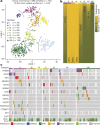
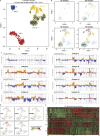
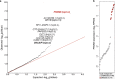
Current therapies for medulloblastoma, a highly malignant childhood brain tumour, impose debilitating effects on the developing child, and highlight the need for molecularly targeted treatments with reduced toxicity. Previous studies have been unable to identify the full spectrum of driver genes and molecular processes that operate in medulloblastoma subgroups. Here we analyse the somatic landscape across 491 sequenced medulloblastoma samples and the molecular heterogeneity among 1,256 epigenetically analysed cases, and identify subgroup-specific driver alterations that include previously undiscovered actionable targets. Driver mutations were confidently assigned to most patients belonging to Group 3 and Group 4 medulloblastoma subgroups, greatly enhancing previous knowledge. New molecular subtypes were differentially enriched for specific driver events, including hotspot in-frame insertions that target KBTBD4 and 'enhancer hijacking' events that activate PRDM6. Thus, the application of integrative genomics to an extensive cohort of clinical samples derived from a single childhood cancer entity revealed a series of cancer genes and biologically relevant subtype diversity that represent attractive therapeutic targets for the treatment of patients with medulloblastoma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

