Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles
- PMID: 28729688
- PMCID: PMC5519541
- DOI: 10.1038/s41467-017-00128-z
Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles
Erratum in
-
Author Correction: Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles.Nat Commun. 2017 Dec 12;8(1):2134. doi: 10.1038/s41467-017-02128-5. Nat Commun. 2017. PMID: 29233980 Free PMC article.
Abstract
Microbial single-cell genomics can be used to provide insights into the metabolic potential, interactions, and evolution of uncultured microorganisms. Here we present WGA-X, a method based on multiple displacement amplification of DNA that utilizes a thermostable mutant of the phi29 polymerase. WGA-X enhances genome recovery from individual microbial cells and viral particles while maintaining ease of use and scalability. The greatest improvements are observed when amplifying high G+C content templates, such as those belonging to the predominant bacteria in agricultural soils. By integrating WGA-X with calibrated index-cell sorting and high-throughput genomic sequencing, we are able to analyze genomic sequences and cell sizes of hundreds of individual, uncultured bacteria, archaea, protists, and viral particles, obtained directly from marine and soil samples, in a single experiment. This approach may find diverse applications in microbiology and in biomedical and forensic studies of humans and other multicellular organisms.Single-cell genomics can be used to study uncultured microorganisms. Here, Stepanauskas et al. present a method combining improved multiple displacement amplification and FACS, to obtain genomic sequences and cell size information from uncultivated microbial cells and viral particles in environmental samples.
Conflict of interest statement
A.L. and T.P. work for Thermo Fisher Scientific, which markets several reagents that were used in this study. R.S., E.A.F., J.B., N.J.P., B.T., J.M.L., E.D.B, J.M.B., M.G.P., B.P.T., C.J.M., and W.K.B. are employees of Bigelow Laboratory for Ocean Sciences, which provides per-fee core facility services in single-cell genomics.
Figures

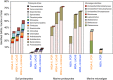
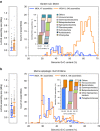

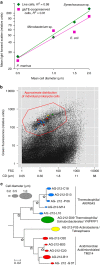
References
-
- Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

