Protein-protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity
- PMID: 28729757
- PMCID: PMC5512853
- DOI: 10.6026/97320630013164
Protein-protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity
Abstract
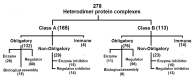
Several catalysis, cellular regulation, immune function, cell wall assembly, transport, signaling and inhibition occur through Protein- Protein Interactions (PPI). This is possible with the formation of specific yet stable protein-protein interfaces. Therefore, it is of interest to understand its molecular principles using structural data in relation to known function. Several interface features have been documented using known X-ray structures of protein complexes since 1975. This has improved our understanding of the interface using structural features such as interface area, binding energy, hydrophobicity, relative hydrophobicity, salt bridges and hydrogen bonds. The strength of binding between two proteins is dependent on interface size (number of residues at the interface) and thus its corresponding interface area. It is known that large interfaces have high binding energy (sum of (van der Waals) vdW, H-bonds, electrostatics). However, the selective role played by each of these energy components and more especially that of vdW is not explicitly known. Therefore, it is important to document their individual role in known protein-protein structural complexes. It is of interest to relate interface size with vdW, H-bonds and electrostatic interactions at the interfaces of protein structural complexes with known function using statistical and multiple linear regression analysis methods to identify the prominent force. We used the manually curated non-redundant dataset of 278 hetero-dimeric protein structural complexes grouped using known functions by Sowmya et al. (2015) to gain additional insight to this phenomenon using a robust inter-atomic non-covalent interaction analyzing tool PPCheck (Anshul and Sowdhamini, 2015). This dataset consists of obligatory (enzymes, regulator, biological assembly), immune and nonobligatory (enzyme and regulator inhibitors) complexes. Results show that the total binding energy is more for large interfaces. However, this is not true for its individual energy factors. Analysis shows that vdW energies contribute to about 75% ± 11% on average among all complexes and it also increases with interface size (r2 ranging from 0.67 to 0.89 with p<0.01) at 95% confidence limit irrespective of molecular function. Thus, vdW is both dominant and proportional at the interface independent of molecular function. Nevertheless, H bond energy contributes to 15% ± 6.5% on average in these complexes. It also moderately increases with interface size (r2 ranging from 0.43 to 0.61 with p<0.01) only among obligatory and immune complexes. Moreover, there is about 11.3% ± 8.7% contribution by electrostatic energy. It increases with interface size specifically among non-obligatory regulator-inhibitors (r2 = 0.44). It is implied that both H-bonds and electrostatics are neither dominant nor proportional at the interface. Nonetheless, their presence cannot be ignored in binding. Therefore, H-bonds and (or) electrostatic energy having specific role for improved stability in complexes is implied. Thus, vdW is common at the interface stabilized further with selective H-bonds and (or) electrostatic interactions at an atomic level in almost all complexes. Comparison of this observation with residue level analysis of the interface is compelling. The role by H-bonds (14.83% ± 6.5% and r2 = 0.61 with p<0.01) among obligatory and electrostatic energy (8.8% ± 4.77% and r2 = 0.63 with p <0.01) among non-obligatory complexes within interfaces (class A) having more non-polar residues than surface is influencing our inference. However, interfaces (class B) having less non-polar residues than surface show 1.5 fold more electrostatic energy on average. The interpretation of the interface using inter-atomic (vdW, H-bonds, electrostatic) interactions combined with inter-residue predominance (class A and class B) in relation to known function is the key to reveal its molecular principles with new challenges.
Keywords: PPI; electrostatics; energy; hydrogen bonds (H-bonds); interface; molecular function; van der Waals (vdW).
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
