Assessment of isomiR Discrimination Using Commercial qPCR Methods
- PMID: 28730153
- PMCID: PMC5514785
- DOI: 10.3390/ncrna3020018
Assessment of isomiR Discrimination Using Commercial qPCR Methods
Abstract
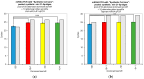
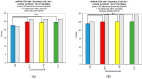
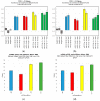
We sought to determine whether commercial quantitative polymerase chain reaction (qPCR) methods are capable of distinguishing isomiRs: variants of mature microRNAs (miRNAs) with sequence endpoint differences. We used two commercially available miRNA qPCR methods to quantify miR-21-5p in both synthetic and real cell contexts. We find that although these miRNA qPCR methods possess high sensitivity for specific sequences, they also pick up background signals from closely related isomiRs, which influences the reliable quantification of individual isomiRs. We conclude that these methods do not possess the requisite specificity for reliable isomiR quantification.
Keywords: isomiR; miRNA; qPCR; short non-coding RNA.
Conflict of interest statement
Conflicts of Interest: The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

