Proteomics analysis reveals a dynamic diurnal pattern of photosynthesis-related pathways in maize leaves
- PMID: 28732011
- PMCID: PMC5521766
- DOI: 10.1371/journal.pone.0180670
Proteomics analysis reveals a dynamic diurnal pattern of photosynthesis-related pathways in maize leaves
Abstract
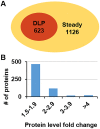
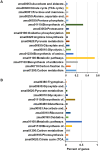
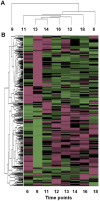
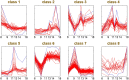
Plant leaves exhibit differentiated patterns of photosynthesis rates under diurnal light regulation. Maize leaves show a single-peak pattern without photoinhibition at midday when the light intensity is maximized. This mechanism contributes to highly efficient photosynthesis in maize leaves. To understand the molecular basis of this process, an isobaric tag for relative and absolute quantitation (iTRAQ)-based proteomics analysis was performed to reveal the dynamic pattern of proteins related to photosynthetic reactions. Steady, single-peak and double-peak protein expression patterns were discovered in maize leaves, and antenna proteins in these leaves displayed a steady pattern. In contrast, the photosystem, carbon fixation and citrate pathways were highly controlled by diurnal light intensity. Most enzymes in the limiting steps of these pathways were major sites of regulation. Thus, maize leaves optimize photosynthesis and carbon fixation outside of light harvesting to adapt to the changes in diurnal light intensity at the protein level.
Conflict of interest statement
Figures
References
-
- Murchie EH, Pinto M, Horton P. Agriculture and the new challenges for photosynthesis research. New Phytologist. 2009;181(3):532–52. doi: 10.1111/j.1469-8137.2008.02705.x - DOI - PubMed
-
- Harmer SL. The circadian system in higher plants. Annual Review of Plant Biology. 2009;60(1):357–77. - PubMed
-
- Zhang Y, Wagle P, Guanter L, Jin C, Xiao X. Phenology and gross primary production of maize croplands from chlorophyll light absorption, solar-induced chlorophyll fluorescence and CO2 flux tower approaches. Jmaterchema. 2015;3(18).
-
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9:R130 Genome Biology. 2008;9(8):: R130. doi: 10.1186/gb-2008-9-8-r130 - DOI - PMC - PubMed
-
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Tong Z, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–3. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

