Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing
- PMID: 28732043
- PMCID: PMC5521745
- DOI: 10.1371/journal.pone.0179880
Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing
Abstract
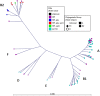
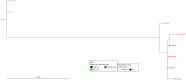
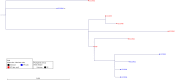
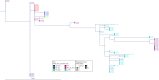
Horizontal gene transfer (HGT) contributes to the evolution of bacteria. All extraintestinal pathogenic Escherichia coli (ExPEC) harbour pathogenicity islands (PAIs), however relatively little is known about the acquisition of these PAIs. Due to these islands, ExPEC have properties to colonize and invade its hosts efficiently. Even though these PAIs are known to be acquired by HGT, only very few PAIs do carry mobilization and transfer genes required for the transmission by HGT. In this study, we apply for the first time next-generation sequencing (NGS) and in silico analyses in combination with in vitro experiments to decipher the mechanisms of PAI acquisition in ExPEC. For this, we investigated three neighbouring E. coli PAIs, namely the high-pathogenicity island (HPI), the pks and the serU island. As these PAIs contain no mobilization and transfer genes, they are immobile and dependent on transfer vehicles. By whole genome sequencing of the entire E. coli reference (ECOR) collection and by applying a phylogenetic approach we could unambiguously demonstrate that these PAIs are transmitted not only vertically, but also horizontally. Furthermore, we could prove in silico that distinct groups of PAIs were transferred "en bloc" in conjunction with the neighbouring chromosomal backbone. We traced this PAI transfer in vitro using an F' plasmid. Different lengths of transferred DNA were exactly detectable in the sequenced transconjugants indicating NGS as a powerful tool for determination of PAI transfer.
Conflict of interest statement
Figures




References
-
- Schneider G, Dobrindt U, Middendorf B, Hochhut B, Szijarto V, Emody L, et al. Mobilisation and remobilisation of a large archetypal pathogenicity island of uropathogenic Escherichia coli in vitro support the role of conjugation for horizontal transfer of genomic islands. BMC Microbiol 2011;11:210 doi: 10.1186/1471-2180-11-210 - DOI - PMC - PubMed
-
- Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, et al. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun 2002. November;70(11):6365–72. doi: 10.1128/IAI.70.11.6365-6372.2002 - DOI - PMC - PubMed
-
- Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, et al. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 2009. January;5(1):e1000257 doi: 10.1371/journal.ppat.1000257 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

