UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses
- PMID: 28732212
- PMCID: PMC5516091
- DOI: 10.1016/j.neo.2017.05.002
UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses
Abstract
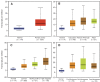
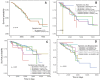
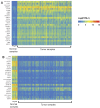
Genomics data from The Cancer Genome Atlas (TCGA) project has led to the comprehensive molecular characterization of multiple cancer types. The large sample numbers in TCGA offer an excellent opportunity to address questions associated with tumo heterogeneity. Exploration of the data by cancer researchers and clinicians is imperative to unearth novel therapeutic/diagnostic biomarkers. Various computational tools have been developed to aid researchers in carrying out specific TCGA data analyses; however there is need for resources to facilitate the study of gene expression variations and survival associations across tumors. Here, we report UALCAN, an easy to use, interactive web-portal to perform to in-depth analyses of TCGA gene expression data. UALCAN uses TCGA level 3 RNA-seq and clinical data from 31 cancer types. The portal's user-friendly features allow to perform: 1) analyze relative expression of a query gene(s) across tumor and normal samples, as well as in various tumor sub-groups based on individual cancer stages, tumor grade, race, body weight or other clinicopathologic features, 2) estimate the effect of gene expression level and clinicopathologic features on patient survival; and 3) identify the top over- and under-expressed (up and down-regulated) genes in individual cancer types. This resource serves as a platform for in silico validation of target genes and for identifying tumor sub-group specific candidate biomarkers. Thus, UALCAN web-portal could be extremely helpful in accelerating cancer research. UALCAN is publicly available at http://ualcan.path.uab.edu.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4(4):346–355. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

