Cutting Edge: CD3 ITAM Diversity Is Required for Optimal TCR Signaling and Thymocyte Development
- PMID: 28733484
- PMCID: PMC5568475
- DOI: 10.4049/jimmunol.1700069
Cutting Edge: CD3 ITAM Diversity Is Required for Optimal TCR Signaling and Thymocyte Development
Abstract
For the αβ or γδTCR chains to integrate extracellular stimuli into the appropriate intracellular cellular response, they must use the 10 ITAMs found within the CD3 subunits (CD3γε, CD3δε, and ζζ) of the TCR signaling complex. However, it remains unclear whether each specific ITAM sequence of the individual subunit (γεδζ) is required for thymocyte development or whether any particular CD3 ITAM motif is sufficient. In this article, we show that mice utilizing a single ITAM sequence (γ, ε, δ, ζa, ζb, or ζc) at each of the 10 ITAM locations exhibit a substantial reduction in thymic cellularity and limited CD4-CD8- (double-negative) to CD4+CD8+ (double-positive) maturation because of low TCR expression and signaling. Together, the data suggest that ITAM sequence diversity is required for optimal TCR signal transduction and subsequent T cell maturation.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures


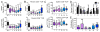
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

