A novel genetic variant of Streptococcus pneumoniae serotype 11A discovered in Fiji
- PMID: 28736074
- PMCID: PMC5869949
- DOI: 10.1016/j.cmi.2017.06.031
A novel genetic variant of Streptococcus pneumoniae serotype 11A discovered in Fiji
Abstract
Objectives: As part of annual cross-sectional Streptococcus pneumoniae carriage surveys in Fiji (2012-2015), we detected pneumococci in over 100 nasopharyngeal swabs that serotyped as '11F-like' by microarray. We examined the genetic basis of this divergence in the 11F-like capsular polysaccharide (cps) locus compared to the reference 11F cps sequence. The impact of this diversity on capsule phenotype, and serotype results using genetic and serologic methods were determined.
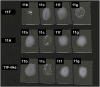
Methods: Genomic DNA from representative 11F-like S. pneumoniae isolates obtained from the nasopharynx of Fijian children was extracted and subject to whole genome sequencing. Genetic and phylogenetic analyses were used to identify genetic changes in the cps locus. Capsular phenotypes were evaluated using the Quellung reaction and latex agglutination.
Results: Compared to published 11F sequences, the wcwC and wcrL genes of the 11F-like cps locus are phylogenetically divergent, and the gct gene contains a single nucleotide insertion within a homopolymeric region. These changes within the DNA sequence of the 11F-like cps locus have modified the antigenic properties of the capsule, such that 11F-like isolates serotype as 11A by Quellung reaction and latex agglutination.
Conclusions: This study demonstrates the ability of molecular serotyping by microarray to identify genetic variants of S. pneumoniae and highlights the potential for discrepant results between phenotypic and genotypic serotyping methods. We propose that 11F-like isolates are not a new serotype but rather are a novel genetic variant of serotype 11A. These findings have implications for invasive pneumococcal disease surveillance as well as studies investigating vaccine impact.
Keywords: Atypical pneumococcal serotype; Genetic variant; Pneumococcal capsule; Pneumococcal serotyping; Pneumococcus; Streptococcus pneumoniae.
Copyright © 2017 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures



References
-
- O'Brien K.L., Wolfson L.J., Watt J.P., Henkle E., Deloria-Knoll M., McCall N. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. - PubMed
-
- Simell B., Auranen K., Käyhty H., Goldblatt D., Dagan R., O'Brien K.L. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. 2012;11:841–855. - PubMed
-
- Bogaert D., de Groot R., Hermans P. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. - PubMed
-
- Diavatopoulos D.A., Short K.R., Price J.T., Wilksch J.J., Brown L.E., Briles D.E. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 2010;24:1789–1798. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

