Channel opening and gating mechanism in AMPA-subtype glutamate receptors
- PMID: 28737760
- PMCID: PMC5743206
- DOI: 10.1038/nature23479
Channel opening and gating mechanism in AMPA-subtype glutamate receptors
Abstract
AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid)-subtype ionotropic glutamate receptors mediate fast excitatory neurotransmission throughout the central nervous system. Gated by the neurotransmitter glutamate, AMPA receptors are critical for synaptic strength, and dysregulation of AMPA receptor-mediated signalling is linked to numerous neurological diseases. Here we use cryo-electron microscopy to solve the structures of AMPA receptor-auxiliary subunit complexes in the apo, antagonist- and agonist-bound states and determine the iris-like mechanism of ion channel opening. The ion channel selectivity filter is formed by the extended portions of the re-entrant M2 loops, while the helical portions of M2 contribute to extensive hydrophobic interfaces between AMPA receptor subunits in the ion channel. We show how the permeation pathway changes upon channel opening and identify conformational changes throughout the entire AMPA receptor that accompany activation and desensitization. Our findings provide a framework for understanding gating across the family of ionotropic glutamate receptors and the role of AMPA receptors in excitatory neurotransmission.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








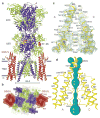

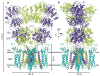


References
-
- Henley JM, Wilkinson KA. Synaptic AMPA receptor composition in development, plasticity and disease. Nature reviews. Neuroscience. 2016;17:337–350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

