Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum
- PMID: 28737826
- PMCID: PMC5548045
- DOI: 10.3892/ijmm.2017.3080
Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum
Abstract
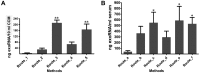
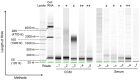
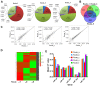
Exosomes are cell-derived vesicles and are abundant in biological fluids; they contain RNA molecules which may serve as potential diagnostic biomarkers in 'precision medicine'. To promote the clinical application of exosomal RNA (exoRNA), many isolation methods must be compared and validated. Exosomes in cell culture medium (CCM) and serum may be isolated using ultracentrifugation (UC), ExoQuick or Total Exosome Isolation Reagent (TEI), and exoRNA may be extracted using TRIzol-LS, SeraMir, Total Exosome RNA Isolation (TER), HiPure Liquid RNA/miRNA kit (HLR), miRNeasy or exoRNeasy. ExoRNA was assessed using NanoDrop, Bioanalyzer 2100, quantitative polymerase chain reaction and high-throughput sequencing. UC showed the lowest recovery of particles, but the highest protein purity for exosome isolation. For isolation of exoRNA, we found that combinations of the TEI and TER methods resulted in high extraction efficiency and purity of small RNA obtained using CCM. High yield and a narrow size distribution pattern of small RNA were shown in exoRNA isolated by exoRNeasy from serum. In RNA profile analysis, the small RNA constituent ratio, miRNA content and amount varied as a result of methodological differences. This study showed that different methods may introduce variations in the concentration, purity and size of exosomes and exoRNA. Herein we discuss the advantages and disadvantages of each method and their application to different materials, therefore providing a reference according to research design.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

