Short-term sleep deprivation leads to decreased systemic redox metabolites and altered epigenetic status
- PMID: 28738082
- PMCID: PMC5524320
- DOI: 10.1371/journal.pone.0181978
Short-term sleep deprivation leads to decreased systemic redox metabolites and altered epigenetic status
Abstract
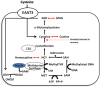
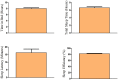
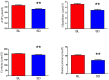
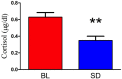
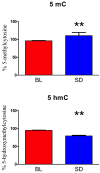
Sleep is critical for repair as well as the rejuvenation processes in the body and many of these functions are regulated via underlying cellular metabolic homeostasis. Changes in sleep pattern are reported to alter such metabolic function resulting in altered disease susceptibility or behavior. Here, we measured the extent to which overnight total sleep deprivation (SD) in young adult humans can influence systemic (plasma-derived) redox-metabolism including the major antioxidant, glutathione as well as DNA methylation levels. Nineteen participants (n = 19, μ age = 21, SD = 3.09) underwent morning testing before and after overnight total SD. Biochemical measures before and after SD revealed that glutathione, ATP, cysteine, and homocysteine levels were significantly reduced following one night of sleep deprivation (all p's < 0.01). Parallel to the well-recognized fact that sleep deprivation (maintaining wakefulness) uses up metabolic reserves, we observed that morning cortisol levels were blunted after sleep deprivation. There were no significant correlations between self-reported or actigraphy-measured sleep and the biochemical measurements, strongly indicating that prior sleep behavior did not have any direct influence on the biochemical measures taken at baseline or after sleep deprivation. Results from the current investigation supports the previous literature implicating the induction of oxidative stress and ATP depletion with sleep deprivation. Furthermore, such altered antioxidant status can also induce downstream epigenetic changes. Although we did not measure the specific genes that were altered under the influence of such sleep deprivation, such epigenetic changes could potentially contribute towards disease predisposition.
Conflict of interest statement
Figures
Similar articles
-
Acute Sleep Loss Induces Tissue-Specific Epigenetic and Transcriptional Alterations to Circadian Clock Genes in Men.J Clin Endocrinol Metab. 2015 Sep;100(9):E1255-61. doi: 10.1210/JC.2015-2284. Epub 2015 Jul 13. J Clin Endocrinol Metab. 2015. PMID: 26168277 Clinical Trial.
-
Spontaneous sleep-wake cycle and sleep deprivation differently induce Bdnf1, Bdnf4 and Bdnf9a DNA methylation and transcripts levels in the basal forebrain and frontal cortex in rats.J Sleep Res. 2015 Apr;24(2):124-30. doi: 10.1111/jsr.12242. Epub 2014 Sep 15. J Sleep Res. 2015. PMID: 25223586
-
How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome.J Sleep Res. 2015 Oct;24(5):476-93. doi: 10.1111/jsr.12307. Epub 2015 Jun 8. J Sleep Res. 2015. PMID: 26059855 Review.
-
Whole blood genome-wide gene expression profile in males after prolonged wakefulness and sleep recovery.Physiol Genomics. 2012 Nov 1;44(21):1003-12. doi: 10.1152/physiolgenomics.00058.2012. Epub 2012 Sep 4. Physiol Genomics. 2012. PMID: 22947657
-
Molecular analysis of sleep.Cold Spring Harb Symp Quant Biol. 2007;72:573-8. doi: 10.1101/sqb.2007.72.054. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419317 Review.
Cited by
-
Redox Balance Correlates with Nutritional Status among Patients with End-Stage Renal Disease Treated with Maintenance Hemodialysis.Oxid Med Cell Longev. 2019 Sep 5;2019:6309465. doi: 10.1155/2019/6309465. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31583040 Free PMC article.
-
The metabolic face of migraine - from pathophysiology to treatment.Nat Rev Neurol. 2019 Nov;15(11):627-643. doi: 10.1038/s41582-019-0255-4. Epub 2019 Oct 4. Nat Rev Neurol. 2019. PMID: 31586135 Review.
-
Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes.Int J Mol Sci. 2022 May 3;23(9):5085. doi: 10.3390/ijms23095085. Int J Mol Sci. 2022. PMID: 35563473 Free PMC article.
-
Influence of Practice Periodization and Sleep Duration on Oxidative Stress in High School Judo Athletes.Sports (Basel). 2023 Aug 30;11(9):163. doi: 10.3390/sports11090163. Sports (Basel). 2023. PMID: 37755840 Free PMC article.
-
Gene expression correlates of advanced epigenetic age and psychopathology in postmortem cortical tissue.Neurobiol Stress. 2021 Jul 29;15:100371. doi: 10.1016/j.ynstr.2021.100371. eCollection 2021 Nov. Neurobiol Stress. 2021. PMID: 34458511 Free PMC article.
References
-
- Knutson KL, Spiegel K, Penev P, Van Cauter E. The Metabolic Consequences of Sleep Deprivation. Sleep Med Rev. 2007;11: 163–178. doi: 10.1016/j.smrv.2007.01.002 - DOI - PMC - PubMed
-
- Villafuerte G, Miguel-Puga A, n, Rodrí M, Guez E, Machado S, et al. Sleep Deprivation and Oxidative Stress in Animal Models: A Systematic Review. Oxid Med Cell Longev. 2015;2015: e234952 doi: 10.1155/2015/234952 - DOI - PMC - PubMed
-
- Gopalakrishnan A, Ji LL, Cirelli C. Sleep deprivation and cellular responses to oxidative stress. Sleep. 2004;27: 27–35. - PubMed
-
- Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol Chem. 2009;390: 191–214. doi: 10.1515/BC.2009.033 - DOI - PMC - PubMed
-
- Trivedi MS, Deth RC. Role of a redox-based methylation switch in mRNA life cycle (pre- and post-transcriptional maturation) and protein turnover: implications in neurological disorders. Front Neurosci. 2012;6: 92 doi: 10.3389/fnins.2012.00092 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

