cGAS surveillance of micronuclei links genome instability to innate immunity
- PMID: 28738408
- PMCID: PMC5870830
- DOI: 10.1038/nature23449
cGAS surveillance of micronuclei links genome instability to innate immunity
Abstract
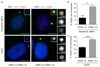
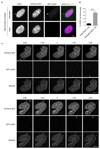
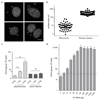
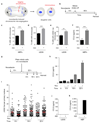
DNA is strictly compartmentalized within the nucleus to prevent autoimmunity; despite this, cyclic GMP-AMP synthase (cGAS), a cytosolic sensor of double-stranded DNA, is activated in autoinflammatory disorders and by DNA damage. Precisely how cellular DNA gains access to the cytoplasm remains to be determined. Here, we report that cGAS localizes to micronuclei arising from genome instability in a mouse model of monogenic autoinflammation, after exogenous DNA damage and spontaneously in human cancer cells. Such micronuclei occur after mis-segregation of DNA during cell division and consist of chromatin surrounded by its own nuclear membrane. Breakdown of the micronuclear envelope, a process associated with chromothripsis, leads to rapid accumulation of cGAS, providing a mechanism by which self-DNA becomes exposed to the cytosol. cGAS is activated by chromatin, and consistent with a mitotic origin, micronuclei formation and the proinflammatory response following DNA damage are cell-cycle dependent. By combining live-cell laser microdissection with single cell transcriptomics, we establish that interferon-stimulated gene expression is induced in micronucleated cells. We therefore conclude that micronuclei represent an important source of immunostimulatory DNA. As micronuclei formed from lagging chromosomes also activate this pathway, recognition of micronuclei by cGAS may act as a cell-intrinsic immune surveillance mechanism that detects a range of neoplasia-inducing processes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




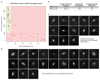
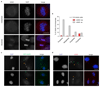


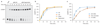
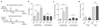

Comment in
-
cGAS Micro-Manages Genotoxic Stress.Immunity. 2017 Oct 17;47(4):616-617. doi: 10.1016/j.immuni.2017.09.020. Immunity. 2017. PMID: 29045895
-
Cell Biology: When Your Own Chromosomes Act like Foreign DNA.Curr Biol. 2017 Nov 20;27(22):R1228-R1231. doi: 10.1016/j.cub.2017.09.043. Curr Biol. 2017. PMID: 29161564
References
-
- Roers A, Hiller B, Hornung V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity. 2016;44:739–754. - PubMed
-
- Hartlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

