CDKL5 localizes at the centrosome and midbody and is required for faithful cell division
- PMID: 28740074
- PMCID: PMC5524905
- DOI: 10.1038/s41598-017-05875-z
CDKL5 localizes at the centrosome and midbody and is required for faithful cell division
Abstract
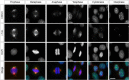
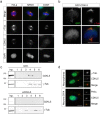
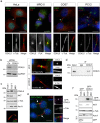
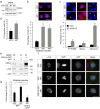
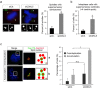
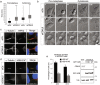
The cyclin-dependent kinase-like 5 (CDKL5) gene has been associated with rare neurodevelopmental disorders characterized by the early onset of seizures and intellectual disability. The CDKL5 protein is widely expressed in most tissues and cells with both nuclear and cytoplasmic localization. In post-mitotic neurons CDKL5 is mainly involved in dendritic arborization, axon outgrowth, and spine formation while in proliferating cells its function is still largely unknown. Here, we report that CDKL5 localizes at the centrosome and at the midbody in proliferating cells. Acute inactivation of CDKL5 by RNA interference (RNAi) leads to multipolar spindle formation, cytokinesis failure and centrosome accumulation. At the molecular level, we observed that, among the several midbody components we analyzed, midbodies of CDKL5-depleted cells were devoid of HIPK2 and its cytokinesis target, the extrachromosomal histone H2B phosphorylated at S14. Of relevance, expression of the phosphomimetic mutant H2B-S14D, which is capable of overcoming cytokinesis failure in HIPK2-defective cells, was sufficient to rescue spindle multipolarity in CDKL5-depleted cells. Taken together, these results highlight a hitherto unknown role of CDKL5 in regulating faithful cell division by guaranteeing proper HIPK2/H2B functions at the midbody.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

