Novel risk genes for systemic lupus erythematosus predicted by random forest classification
- PMID: 28740209
- PMCID: PMC5524838
- DOI: 10.1038/s41598-017-06516-1
Novel risk genes for systemic lupus erythematosus predicted by random forest classification
Abstract
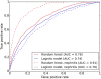
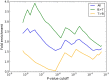
Genome-wide association studies have identified risk loci for SLE, but a large proportion of the genetic contribution to SLE still remains unexplained. To detect novel risk genes, and to predict an individual's SLE risk we designed a random forest classifier using SNP genotype data generated on the "Immunochip" from 1,160 patients with SLE and 2,711 controls. Using gene importance scores defined by the random forest classifier, we identified 15 potential novel risk genes for SLE. Of them 12 are associated with other autoimmune diseases than SLE, whereas three genes (ZNF804A, CDK1, and MANF) have not previously been associated with autoimmunity. Random forest classification also allowed prediction of patients at risk for lupus nephritis with an area under the curve of 0.94. By allele-specific gene expression analysis we detected cis-regulatory SNPs that affect the expression levels of six of the top 40 genes designed by the random forest analysis, indicating a regulatory role for the identified risk variants. The 40 top genes from the prediction were overrepresented for differential expression in B and T cells according to RNA-sequencing of samples from five healthy donors, with more frequent over-expression in B cells compared to T cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

