Structure Modeling of Human Tyrosyl-DNA Phosphodiesterase 1 and Screening for Its Inhibitors
- PMID: 28740727
- PMCID: PMC5509001
Structure Modeling of Human Tyrosyl-DNA Phosphodiesterase 1 and Screening for Its Inhibitors
Abstract
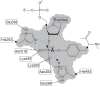
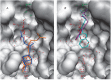
The DNA repair enzyme tyrosyl-DNA phosphodiesterase 1 (Tdp1) represents a potential molecular target for anticancer therapy. A human Tdp1 model has been constructed using the methods of quantum and molecular mechanics, taking into account the ionization states of the amino acid residues in the active site and their interactions with the substrate and competitive inhibitors. The oligonucleotide- and phosphotyrosine-binding cavities important for the inhibitor design have been identified in the enzyme's active site. The developed molecular model allowed us to uncover new Tdp1 inhibitors whose sulfo group is capable of occupying the position of the 3'-phosphate group of the substrate and forming hydrogen bonds with Lys265, Lys495, and other amino acid residues in the phosphotyrosine binding site.
Keywords: docking; inhibitor; molecular modeling; tyrosyl-DNA phosphodiesterase 1.
Figures



References
-
- Champoux J.J.. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Wang J.C.. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. - PubMed
-
- Pommier Y.. Nat. Rev. Cancer. 2006;6:789–802. - PubMed
-
- Lebedeva N., Rechkunova N., Boiteux S., Lavrik O.. IUBMB Life. 2008;60:130–134. - PubMed
-
- Pommier Y., Redon C., Rao V.A., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G.. Mutat. Res. 2003;532:173–203. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
