An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome
- PMID: 28741611
- PMCID: PMC5589491
- DOI: 10.1038/nsmb.3439
An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome
Abstract
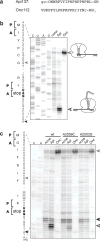
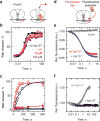
Many antibiotics stop bacterial growth by inhibiting different steps of protein synthesis. However, no specific inhibitors of translation termination are known. Proline-rich antimicrobial peptides, a component of the antibacterial defense system of multicellular organisms, interfere with bacterial growth by inhibiting translation. Here we show that Api137, a derivative of the insect-produced antimicrobial peptide apidaecin, arrests terminating ribosomes using a unique mechanism of action. Api137 binds to the Escherichia coli ribosome and traps release factor (RF) RF1 or RF2 subsequent to the release of the nascent polypeptide chain. A high-resolution cryo-EM structure of the ribosome complexed with RF1 and Api137 reveals the molecular interactions that lead to RF trapping. Api137-mediated depletion of the cellular pool of free release factors causes the majority of ribosomes to stall at stop codons before polypeptide release, thereby resulting in a global shutdown of translation termination.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

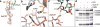

Similar articles
-
Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.Nat Commun. 2024 May 10;15(1):3945. doi: 10.1038/s41467-024-48027-4. Nat Commun. 2024. PMID: 38730238 Free PMC article.
-
Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.Nat Commun. 2018 Aug 3;9(1):3053. doi: 10.1038/s41467-018-05465-1. Nat Commun. 2018. PMID: 30076302 Free PMC article.
-
Mechanistic insights into the alternative translation termination by ArfA and RF2.Nature. 2017 Jan 26;541(7638):550-553. doi: 10.1038/nature20822. Epub 2016 Dec 1. Nature. 2017. PMID: 27906160
-
Molecular recognition and catalysis in translation termination complexes.Trends Biochem Sci. 2011 May;36(5):282-92. doi: 10.1016/j.tibs.2011.02.001. Epub 2011 Mar 17. Trends Biochem Sci. 2011. PMID: 21420300 Review.
-
Intracellular Antimicrobial Peptides Targeting the Protein Synthesis Machinery.Adv Exp Med Biol. 2019;1117:73-89. doi: 10.1007/978-981-13-3588-4_6. Adv Exp Med Biol. 2019. PMID: 30980354 Review.
Cited by
-
Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.Nat Chem. 2023 Jan;15(1):143-153. doi: 10.1038/s41557-022-01073-1. Epub 2022 Oct 31. Nat Chem. 2023. PMID: 36316410 Free PMC article.
-
Unveiling mechanisms of antimicrobial peptide: Actions beyond the membranes disruption.Heliyon. 2024 Sep 20;10(19):e38079. doi: 10.1016/j.heliyon.2024.e38079. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39386776 Free PMC article. Review.
-
Protegrin-1 cytotoxicity towards mammalian cells positively correlates with the magnitude of conformational changes of the unfolded form upon cell interaction.Sci Rep. 2019 Aug 9;9(1):11569. doi: 10.1038/s41598-019-47955-2. Sci Rep. 2019. PMID: 31399625 Free PMC article.
-
Genes within Genes in Bacterial Genomes.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0020-2018. doi: 10.1128/microbiolspec.RWR-0020-2018. Microbiol Spectr. 2018. PMID: 30003865 Free PMC article. Review.
-
The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.Nat Chem Biol. 2024 Jul;20(7):877-884. doi: 10.1038/s41589-023-01521-0. Epub 2024 Jan 3. Nat Chem Biol. 2024. PMID: 38172604 Free PMC article.
References
-
- Bremer H, Dennis P. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. Vol. 2. ASM Press; 1996. pp. 1553–1569. Ch. 97.
-
- Shi X, Joseph S. Mechanism of translation termination: RF1 dissociation follows dissociation of RF3 from the ribosome. Biochemistry. 2016;55:6344–6354. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

