Structural origins of clustered protocadherin-mediated neuronal barcoding
- PMID: 28743640
- PMCID: PMC5582985
- DOI: 10.1016/j.semcdb.2017.07.023
Structural origins of clustered protocadherin-mediated neuronal barcoding
Abstract
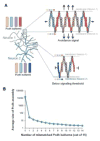
Clustered protocadherins mediate neuronal self-recognition and non-self discrimination-neuronal "barcoding"-which underpin neuronal self-avoidance in vertebrate neurons. Recent structural, biophysical, computational, and cell-based studies on protocadherin structure and function have led to a compelling molecular model for the barcoding mechanism. Protocadherin isoforms assemble into promiscuous cis-dimeric recognition units and mediate cell-cell recognition through homophilic trans-interactions. Each recognition unit is composed of two arms extending from the membrane proximal EC6 domains. A cis-dimeric recognition unit with each arm coding adhesive trans homophilic specificity can generate a zipper-like assembly that in turn suggests a chain termination mechanism for self-vs-non-self-discrimination among vertebrate neurons.
Keywords: Cell-cell recognition; Clustered protocadherins; Crystal structure; Neuronal self-avoidance; Protein interaction specificity.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures






References
-
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

