Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica
- PMID: 28744371
- PMCID: PMC5518176
- DOI: 10.1093/ve/vex017
Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica
Abstract
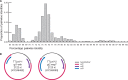
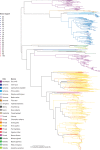
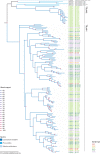
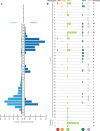
The viruses circulating among Antarctic wildlife remain largely unknown. In an effort to identify viruses associated with Weddell seals (Leptonychotes weddellii) inhabiting the Ross Sea, vaginal and nasal swabs, and faecal samples were collected between November 2014 and February 2015. In addition, a Weddell seal kidney and South Polar skua (Stercorarius maccormicki) faeces were opportunistically sampled. Using high throughput sequencing, we identified and recovered 152 anellovirus genomes that share 63-70% genome-wide identities with other pinniped anelloviruses. Genome-wide pairwise comparisons coupled with phylogenetic analysis revealed two novel anellovirus species, tentatively named torque teno Leptonychotes weddellii virus (TTLwV) -1 and -2. TTLwV-1 (n = 133, genomes encompassing 40 genotypes) is highly recombinant, whereas TTLwV-2 (n = 19, genomes encompassing three genotypes) is relatively less recombinant. This study documents ubiquitous TTLwVs among Weddell seals in Antarctica with frequent co-infection by multiple genotypes, however, the role these anelloviruses play in seal health remains unknown.
Keywords: Anelloviridae; Antarctica; Ross Sea; South Polar skua; Weddell seal.
Figures

References
-
- Al-Moslih M. I., Perkins H., Hu Y. W. (2007) ‘Genetic Relationship of Torque Teno Virus (TTV) between Humans and Camels in United Arab Emirates (UAE)’, Journal of Medical Virology, 79: 188–91. - PubMed
-
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. - PubMed
-
- Austin F. J., Webster R. G. (1993) ‘Evidence of Ortho- and Paramoxyoviruses in Fauana from Antarctica’, Journal of Wildlife Diseases, 29: 568–71. - PubMed
-
- Biagini P. (2009) ‘Classification of TTV and Related Viruses (Anelloviruses)’, in Villiers E.-M., Hausen H. (eds.) TT Viruses, pp. 21–33. Springer Berlin Heidelberg. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

