The Distribution of Miniature Impala Elements and SIX Genes in the Fusarium Genus is Suggestive of Horizontal Gene Transfer
- PMID: 28744785
- PMCID: PMC5579170
- DOI: 10.1007/s00239-017-9801-0
The Distribution of Miniature Impala Elements and SIX Genes in the Fusarium Genus is Suggestive of Horizontal Gene Transfer
Abstract
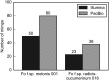
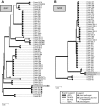
The mimp family of miniature inverted-repeat transposable elements was previously found only in genomes of Fusarium oxysporum and is contextually associated with virulence genes in this species. Through extensive comparative analysis of 83 F. oxysporum and 52 other Fusarium genomes, we uncovered the distribution of different mimp families throughout the genus. We show that (i) mimps are not exclusive to F. oxysporum; (ii) pathogenic isolates generally possess more mimps than non-pathogenic strains and (iii) two isolates of F. hostae and one F. proliferatum isolate display evidence for horizontal transfer of genetic material to or from F. oxysporum. Multiple instances of mimp elements identical to F. oxysporum mimps were encountered in the genomes of these isolates. Moreover, homologs of effector genes (SIX1, 2, 6, 7, 11 and FomAVR2) were discovered here, several with very high (97-100%) pairwise nucleotide sequence identity scores. These three strains were isolated from infected flower bulbs (Hyacinthus and Lilium spp.). Their ancestors may thus have lived in close proximity to pathogenic strains of F. oxysporum f. sp. hyacinthi and f. sp. lilii. The Fo f. sp. lycopersici SIX2 effector gene was found to be widely distributed (15/18 isolates) throughout the F. fujikuroi species complex, exhibiting a predominantly vertical inheritance pattern. These findings shed light on the potential evolutionary mechanism underlying plant-pathogenicity in Fusarium and show that interspecies horizontal gene transfer may have occurred.
Keywords: Comparative genomics; Horizontal gene transfer; Inverted repeat; MITE; Transposable elements; mimp.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Baayen RP, Förch MG, Waalwijk C, et al. Pathogenic, genetic and molecular characterisation of Fusarium oxysporum f. sp. lilii. Eur J Plant Pathol. 1998;104:887–894. doi: 10.1023/A:1008676729143. - DOI
-
- Breeuwsma SJ, De Boer M (2004) Fusarium in bloembolgewassen: detectiemethoden en vruchtwisselingsproblematiek. PPO Bloembollen en Bomen, PPO project 320689 (in Dutch)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

