Stimulation of functional neuronal regeneration from Müller glia in adult mice
- PMID: 28746305
- PMCID: PMC5991837
- DOI: 10.1038/nature23283
Stimulation of functional neuronal regeneration from Müller glia in adult mice
Abstract
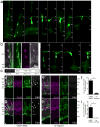
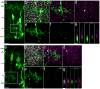
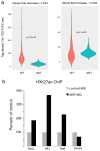
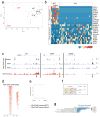
Many retinal diseases lead to the loss of retinal neurons and cause visual impairment. The adult mammalian retina has little capacity for regeneration. By contrast, teleost fish functionally regenerate their retina following injury, and Müller glia (MG) are the source of regenerated neurons. The proneural transcription factor Ascl1 is upregulated in MG after retinal damage in zebrafish and is necessary for regeneration. Although Ascl1 is not expressed in mammalian MG after injury, forced expression of Ascl1 in mouse MG induces a neurogenic state in vitro and in vivo after NMDA (N-methyl-d-aspartate) damage in young mice. However, by postnatal day 16, mouse MG lose neurogenic capacity, despite Ascl1 overexpression. Loss of neurogenic capacity in mature MG is accompanied by reduced chromatin accessibility, suggesting that epigenetic factors limit regeneration. Here we show that MG-specific overexpression of Ascl1, together with a histone deacetylase inhibitor, enables adult mice to generate neurons from MG after retinal injury. The MG-derived neurons express markers of inner retinal neurons, synapse with host retinal neurons, and respond to light. Using an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), we show that the histone deacetylase inhibitor promotes accessibility at key gene loci in the MG, and allows more effective reprogramming. Our results thus provide a new approach for the treatment of blinding retinal diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








References
-
- Fischer AJ, Reh TA. Müller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247–252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

