The Alzheimer's disease-protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool
- PMID: 28747436
- PMCID: PMC5602391
- DOI: 10.1074/jbc.M117.799346
The Alzheimer's disease-protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool
Abstract
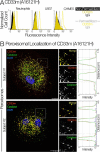
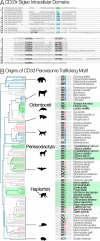
The immunomodulatory receptor Siglec-3/CD33 influences risk for late-onset Alzheimer's disease (LOAD), an apparently human-specific post-reproductive disease. CD33 generates two splice variants: a full-length CD33M transcript produced primarily by the "LOAD-risk" allele and a shorter CD33m isoform lacking the sialic acid-binding domain produced primarily from the "LOAD-protective" allele. An SNP that modulates CD33 splicing to favor CD33m is associated with enhanced microglial activity. Individuals expressing more protective isoform accumulate less brain β-amyloid and have a lower LOAD risk. How the CD33m isoform increases β-amyloid clearance remains unknown. We report that the protection by the CD33m isoform may not be conferred by what it does but, rather, from what it cannot do. Analysis of blood neutrophils and monocytes and a microglial cell line revealed that unlike CD33M, the CD33m isoform does not localize to cell surfaces; instead, it accumulates in peroxisomes. Cell stimulation and activation did not mobilize CD33m to the surface. Thus, the CD33m isoform may neither interact directly with amyloid plaques nor engage in cell-surface signaling. Rather, production and localization of CD33m in peroxisomes is a way of diminishing the amount of CD33M and enhancing β-amyloid clearance. We confirmed intracellular localization by generating a CD33m-specific monoclonal antibody. Of note, CD33 is the only Siglec with a peroxisome-targeting sequence, and this motif emerged by convergent evolution in toothed whales, the only other mammals with a prolonged post-reproductive lifespan. The CD33 allele that protects post-reproductive individuals from LOAD may have evolved by adaptive loss-of-function, an example of the less-is-more hypothesis.
Keywords: Alzheimer disease; CD33; Siglec-3; alternative splicing; intracellular trafficking; peroxisome; sialic acid.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Crocker P. R., Paulson J. C., and Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 - PubMed
-
- Schwarz F., Fong J. J., and Varki A. (2015) Human-specific evolutionary changes in the biology of siglecs. Adv. Exp. Med. Biol. 842, 1–16 - PubMed
-
- Varki A., and Angata T. (2006) Siglecs: the major subfamily of I-type lectins. Glycobiology 16, 1R–27R - PubMed
-
- Walter R. B., Raden B. W., Zeng R., Häusermann P., Bernstein I. D., and Cooper J. A. (2008) ITIM-dependent endocytosis of CD33-related Siglecs: role of intracellular domain, tyrosine phosphorylation, and the tyrosine phosphatases, Shp1 and Shp2. J. Leukoc. Biol. 83, 200–211 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

