Panels of HIV-1 Subtype C Env Reference Strains for Standardized Neutralization Assessments
- PMID: 28747500
- PMCID: PMC5599761
- DOI: 10.1128/JVI.00991-17
Panels of HIV-1 Subtype C Env Reference Strains for Standardized Neutralization Assessments
Erratum in
-
Correction for Hraber et al., "Panels of HIV-1 Subtype C Env Reference Strains for Standardized Neutralization Assessments".J Virol. 2017 Dec 14;92(1):e01817-17. doi: 10.1128/JVI.01817-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29242255 Free PMC article. No abstract available.
Abstract
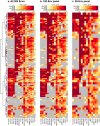
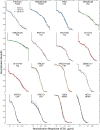
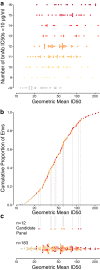
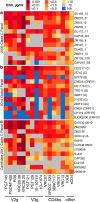
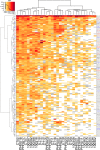
In the search for effective immunologic interventions to prevent and treat HIV-1 infection, standardized reference reagents are a cost-effective way to maintain robustness and reproducibility among immunological assays. To support planned and ongoing studies where clade C predominates, here we describe three virus panels, chosen from 200 well-characterized clade C envelope (Env)-pseudotyped viruses from early infection. All 200 Envs were expressed as a single round of replication pseudoviruses and were tested to quantify neutralization titers by 16 broadly neutralizing antibodies (bnAbs) and sera from 30 subjects with chronic clade C infections. We selected large panels of 50 and 100 Envs either to characterize cross-reactive breadth for sera identified as having potent neutralization activity based on initial screening or to evaluate neutralization magnitude-breadth distributions of newly isolated antibodies. We identified these panels by downselection after hierarchical clustering of bnAb neutralization titers. The resulting panels represent the diversity of neutralization profiles throughout the range of virus sensitivities identified in the original panel of 200 viruses. A small 12-Env panel was chosen to screen sera from vaccine trials or natural-infection studies for neutralization responses. We considered panels selected by previously described methods but favored a computationally informed method that enabled selection of viruses representing diverse neutralization sensitivity patterns, given that we do not a priori know what the neutralization-response profile of vaccine sera will be relative to that of sera from infected individuals. The resulting 12-Env panel complements existing panels. Use of standardized panels enables direct comparisons of data from different trials and study sites testing HIV-1 clade C-specific products.IMPORTANCE HIV-1 group M includes nine clades and many recombinants. Clade C is the most common lineage, responsible for roughly half of current HIV-1 infections, and is a focus for vaccine design and testing. Standard reference reagents, particularly virus panels to study neutralization by antibodies, are crucial for developing cost-effective and yet rigorous and reproducible assays against diverse examples of this variable virus. We developed clade C-specific panels for use as standardized reagents to monitor complex polyclonal sera for neutralization activity and to characterize the potency and breadth of cross-reactive neutralization by monoclonal antibodies, whether engineered or isolated from infected individuals. We chose from 200 southern African, clade C envelope-pseudotyped viruses with neutralization titers against 16 broadly neutralizing antibodies and 30 sera from chronic clade C infections. We selected panels to represent the diversity of bnAb neutralization profiles and Env neutralization sensitivities. Use of standard virus panels can facilitate comparison of results across studies and sites.
Keywords: assay standardization; clinical trials; human immunodeficiency virus; immunoserology; neutralizing antibodies; vaccines.
Copyright © 2017 Hraber et al.
Figures

References
-
- Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol 79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. - DOI - PMC - PubMed
-
- Iversen AK, Learn GH, Skinhoj P, Mullins JI, McMichael AJ, Rambaut A. 2005. Preferential detection of HIV subtype C′ over subtype A in cervical cells from a dually infected woman. AIDS 19:990–993. doi: 10.1097/01.aids.0000171418.91786.ad. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

