Identification of Vaccinia Virus Replisome and Transcriptome Proteins by Isolation of Proteins on Nascent DNA Coupled with Mass Spectrometry
- PMID: 28747503
- PMCID: PMC5599757
- DOI: 10.1128/JVI.01015-17
Identification of Vaccinia Virus Replisome and Transcriptome Proteins by Isolation of Proteins on Nascent DNA Coupled with Mass Spectrometry
Abstract
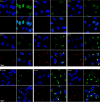
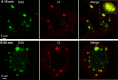
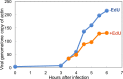
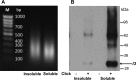
Poxviruses replicate within the cytoplasm and encode proteins for DNA and mRNA synthesis. To investigate poxvirus replication and transcription from a new perspective, we incorporated 5-ethynyl-2'-deoxyuridine (EdU) into nascent DNA in cells infected with vaccinia virus (VACV). The EdU-labeled DNA was conjugated to fluor- or biotin-azide and visualized by confocal, superresolution, and transmission electron microscopy. Nuclear labeling decreased dramatically after infection, accompanied by intense labeling of cytoplasmic foci. The nascent DNA colocalized with the VACV single-stranded DNA binding protein I3 in multiple puncta throughout the interior of factories, which were surrounded by endoplasmic reticulum. Complexes containing EdU-biotin-labeled DNA cross-linked to proteins were captured on streptavidin beads. After elution and proteolysis, the peptides were analyzed by mass spectrometry to identify proteins associated with nascent DNA. The known viral replication proteins, a telomere binding protein, and a protein kinase were associated with nascent DNA, as were the DNA-dependent RNA polymerase and intermediate- and late-stage transcription initiation and elongation factors, plus the capping and methylating enzymes. These results suggested that the replicating pool of DNA is transcribed and that few if any additional viral proteins directly engaged in replication and transcription remain to be discovered. Among the host proteins identified by mass spectrometry, topoisomerases IIα and IIβ and PCNA were noteworthy. The association of the topoisomerases with nascent DNA was dependent on expression of the viral DNA ligase, in accord with previous proteomic studies. Further investigations are needed to determine possible roles for PCNA and other host proteins detected.IMPORTANCE Poxviruses, unlike many well-characterized animal DNA viruses, replicate entirely within the cytoplasm of animal cells, raising questions regarding the relative roles of viral and host proteins. We adapted newly developed procedures for click chemistry and iPOND (
Keywords: DNA binding proteins; DNA replication; iPOND; poxvirus; transcription factors; vaccinia virus.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Nonredundant roles of topoisomerase 2α and 2β in the cytosolic replication of vaccinia virus.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf566. doi: 10.1093/nar/gkaf566. Nucleic Acids Res. 2025. PMID: 40557872 Free PMC article.
-
Identification of Poxvirus Genome Uncoating and DNA Replication Factors with Mutually Redundant Roles.J Virol. 2018 Mar 14;92(7):e02152-17. doi: 10.1128/JVI.02152-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343579 Free PMC article.
-
Identification and characterization of O-GlcNAc modifications of a conserved orthopoxvirus core protein.J Virol. 2025 Jun 17;99(6):e0005825. doi: 10.1128/jvi.00058-25. Epub 2025 May 23. J Virol. 2025. PMID: 40407344 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
Nonredundant roles of topoisomerase 2α and 2β in the cytosolic replication of vaccinia virus.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf566. doi: 10.1093/nar/gkaf566. Nucleic Acids Res. 2025. PMID: 40557872 Free PMC article.
-
Strategies for Success. Viral Infections and Membraneless Organelles.Front Cell Infect Microbiol. 2019 Oct 11;9:336. doi: 10.3389/fcimb.2019.00336. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31681621 Free PMC article. Review.
-
Recent trends in click chemistry as a promising technology for virus-related research.Virus Res. 2018 Sep 2;256:21-28. doi: 10.1016/j.virusres.2018.08.003. Epub 2018 Aug 3. Virus Res. 2018. PMID: 30081058 Free PMC article. Review.
-
Human FAM111A inhibits vaccinia virus replication by degrading viral protein I3 and is antagonized by poxvirus host range factor SPI-1.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2304242120. doi: 10.1073/pnas.2304242120. Epub 2023 Aug 22. Proc Natl Acad Sci U S A. 2023. PMID: 37607234 Free PMC article.
-
Vaccinia Virus C9 Ankyrin Repeat/F-Box Protein Is a Newly Identified Antagonist of the Type I Interferon-Induced Antiviral State.J Virol. 2018 Apr 13;92(9):e00053-18. doi: 10.1128/JVI.00053-18. Print 2018 May 1. J Virol. 2018. PMID: 29444943 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

