Targeted Genome Sequencing Reveals Varicella-Zoster Virus Open Reading Frame 12 Deletion
- PMID: 28747504
- PMCID: PMC5625521
- DOI: 10.1128/JVI.01141-17
Targeted Genome Sequencing Reveals Varicella-Zoster Virus Open Reading Frame 12 Deletion
Abstract
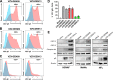
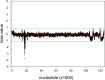
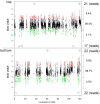
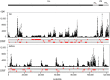
The neurotropic herpesvirus varicella-zoster virus (VZV) establishes a lifelong latent infection in humans following primary infection. The low abundance of VZV nucleic acids in human neurons has hindered an understanding of the mechanisms that regulate viral gene transcription during latency. To overcome this critical barrier, we optimized a targeted capture protocol to enrich VZV DNA and cDNA prior to whole-genome/transcriptome sequence analysis. Since the VZV genome is remarkably stable, it was surprising to detect that VZV32, a VZV laboratory strain with no discernible growth defect in tissue culture, contained a 2,158-bp deletion in open reading frame (ORF) 12. Consequently, ORF 12 and 13 protein expression was abolished and Akt phosphorylation was inhibited. The discovery of the ORF 12 deletion, revealed through targeted genome sequencing analysis, points to the need to authenticate the VZV genome when the virus is propagated in tissue culture.IMPORTANCE Viruses isolated from clinical samples often undergo genetic modifications when cultured in the laboratory. Historically, VZV is among the most genetically stable herpesviruses, a notion supported by more than 60 complete genome sequences from multiple isolates and following multiple in vitro passages. However, application of enrichment protocols to targeted genome sequencing revealed the unexpected deletion of a significant portion of VZV ORF 12 following propagation in cultured human fibroblast cells. While the enrichment protocol did not introduce bias in either the virus genome or transcriptome, the findings indicate the need for authentication of VZV by sequencing when the virus is propagated in tissue culture.
Keywords: ORF 12; VZV; deletion; genome; transcriptome.
Copyright © 2017 American Society for Microbiology.
Figures
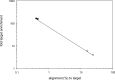
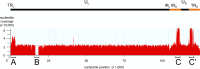
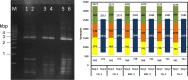
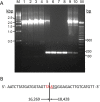



Similar articles
-
Sequence analysis of the leftward end of simian varicella virus (EcoRI-I fragment) reveals the presence of an 8-bp repeat flanking the unique long segment and an 881-bp open-reading frame that is absent in the varicella zoster virus genome.Virology. 2000 Sep 1;274(2):420-8. doi: 10.1006/viro.2000.0465. Virology. 2000. PMID: 10964784
-
Decoding the Architecture of the Varicella-Zoster Virus Transcriptome.mBio. 2020 Oct 6;11(5):e01568-20. doi: 10.1128/mBio.01568-20. mBio. 2020. PMID: 33024035 Free PMC article.
-
Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells.J Virol. 2000 Dec;74(23):11311-21. doi: 10.1128/jvi.74.23.11311-11321.2000. J Virol. 2000. PMID: 11070031 Free PMC article.
-
Molecular characterization of varicella zoster virus in latently infected human ganglia: physical state and abundance of VZV DNA, Quantitation of viral transcripts and detection of VZV-specific proteins.Curr Top Microbiol Immunol. 2010;342:229-41. doi: 10.1007/82_2009_2. Curr Top Microbiol Immunol. 2010. PMID: 20186615 Free PMC article. Review.
-
Latency and reactivation of varicella zoster virus infections.Scand J Infect Dis Suppl. 1996;100:46-50. Scand J Infect Dis Suppl. 1996. PMID: 9163025 Review.
Cited by
-
Uncovering Bifidobacteria via Targeted Sequencing of the Mammalian Gut Microbiota.Microorganisms. 2019 Nov 6;7(11):535. doi: 10.3390/microorganisms7110535. Microorganisms. 2019. PMID: 31698863 Free PMC article.
-
Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus.BMC Genomics. 2018 Dec 4;19(1):873. doi: 10.1186/s12864-018-5267-8. BMC Genomics. 2018. PMID: 30514211 Free PMC article.
-
Hybrid-Capture Target Enrichment in Human Pathogens: Identification, Evolution, Biosurveillance, and Genomic Epidemiology.Pathogens. 2024 Mar 23;13(4):275. doi: 10.3390/pathogens13040275. Pathogens. 2024. PMID: 38668230 Free PMC article. Review.
-
Ultrasensitive Capture of Human Herpes Simplex Virus Genomes Directly from Clinical Samples Reveals Extraordinarily Limited Evolution in Cell Culture.mSphere. 2018 Jun 13;3(3):e00283-18. doi: 10.1128/mSphereDirect.00283-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29898986 Free PMC article.
-
Genetic insights into the dark matter of the mammalian gut microbiota through targeted genome reconstruction.Environ Microbiol. 2021 Jun;23(6):3294-3305. doi: 10.1111/1462-2920.15559. Epub 2021 May 19. Environ Microbiol. 2021. PMID: 33973321 Free PMC article.
References
-
- Weinberg A, Levin MJ. 2010. VZV T cell-mediated immunity. Curr Top Microbiol Immunol 342:341–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

