A single amino acid substitution confers B-cell clonogenic activity to the HIV-1 matrix protein p17
- PMID: 28747658
- PMCID: PMC5529431
- DOI: 10.1038/s41598-017-06848-y
A single amino acid substitution confers B-cell clonogenic activity to the HIV-1 matrix protein p17
Abstract
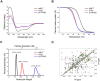
Recent data highlight the presence, in HIV-1-seropositive patients with lymphoma, of p17 variants (vp17s) endowed with B-cell clonogenicity, suggesting a role of vp17s in lymphomagenesis. We investigated the mechanisms responsible for the functional disparity on B cells between a wild-type p17 (refp17) and a vp17 named S75X. Here, we show that a single Arginine (R) to Glycine (G) mutation at position 76 in the refp17 backbone (p17R76G), as in the S75X variant, is per se sufficient to confer a B-cell clonogenic potential to the viral protein and modulate, through activation of the PTEN/PI3K/Akt signaling pathway, different molecules involved in apoptosis inhibition (CASP-9, CASP-7, DFF-45, NPM, YWHAZ, Src, PAX2, MAPK8), cell cycle promotion and cancer progression (CDK1, CDK2, CDK8, CHEK1, CHEK2, GSK-3 beta, NPM, PAK1, PP2C-alpha). Moreover, the only R to G mutation at position 76 was found to strongly impact on protein folding and oligomerization by altering the hydrogen bond network. This generates a conformational shift in the p17 R76G mutant which enables a functional epitope(s), masked in refp17, to elicit B-cell growth-promoting signals after its interaction with a still unknown receptor(s). Our findings offer new opportunities to understand the molecular mechanisms accounting for the B-cell growth-promoting activity of vp17s.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

