4-dimensional functional profiling in the convulsant-treated larval zebrafish brain
- PMID: 28747660
- PMCID: PMC5529444
- DOI: 10.1038/s41598-017-06646-6
4-dimensional functional profiling in the convulsant-treated larval zebrafish brain
Erratum in
-
Author Correction: 4-dimensional functional profiling in the convulsant-treated larval zebrafish brain.Sci Rep. 2018 Oct 23;8(1):15903. doi: 10.1038/s41598-018-32731-5. Sci Rep. 2018. PMID: 30349014 Free PMC article.
Abstract
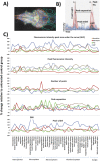
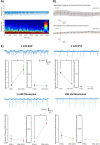
Functional neuroimaging, using genetically-encoded Ca2+ sensors in larval zebrafish, offers a powerful combination of high spatiotemporal resolution and higher vertebrate relevance for quantitative neuropharmacological profiling. Here we use zebrafish larvae with pan-neuronal expression of GCaMP6s, combined with light sheet microscopy and a novel image processing pipeline, for the 4D profiling of chemoconvulsant action in multiple brain regions. In untreated larvae, regions associated with autonomic functionality, sensory processing and stress-responsiveness, consistently exhibited elevated spontaneous activity. The application of drugs targeting different convulsant mechanisms (4-Aminopyridine, Pentylenetetrazole, Pilocarpine and Strychnine) resulted in distinct spatiotemporal patterns of activity. These activity patterns showed some interesting parallels with what is known of the distribution of their respective molecular targets, but crucially also revealed system-wide neural circuit responses to stimulation or suppression. Drug concentration-response curves of neural activity were identified in a number of anatomically-defined zebrafish brain regions, and in vivo larval electrophysiology, also conducted in 4dpf larvae, provided additional measures of neural activity. Our quantification of network-wide chemoconvulsant drug activity in the whole zebrafish brain illustrates the power of this approach for neuropharmacological profiling in applications ranging from accelerating studies of drug safety and efficacy, to identifying pharmacologically-altered networks in zebrafish models of human neurological disorders.
Conflict of interest statement
S.F.O and W.S.R. are employees of AstraZeneca P.L.C. and W.S.R. and M.J.H. are shareholders of AstraZeneca P.L.C. The authors declare no other competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

