Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors
- PMID: 28749946
- PMCID: PMC5531551
- DOI: 10.1371/journal.pone.0179726
Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors
Abstract
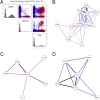
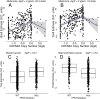
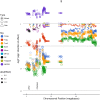
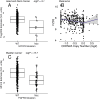
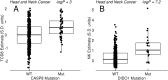
Therapeutic options for the treatment of an increasing variety of cancers have been expanded by the introduction of a new class of drugs, commonly referred to as checkpoint blocking agents, that target the host immune system to positively modulate anti-tumor immune response. Although efficacy of these agents has been linked to a pre-existing level of tumor immune infiltrate, it remains unclear why some patients exhibit deep and durable responses to these agents while others do not benefit. To examine the influence of tumor genetics on tumor immune state, we interrogated the relationship between somatic mutation and copy number alteration with infiltration levels of 7 immune cell types across 40 tumor cohorts in The Cancer Genome Atlas. Levels of cytotoxic T, regulatory T, total T, natural killer, and B cells, as well as monocytes and M2 macrophages, were estimated using a novel set of transcriptional signatures that were designed to resist interference from the cellular heterogeneity of tumors. Tumor mutational load and estimates of tumor purity were included in our association models to adjust for biases in multi-modal genomic data. Copy number alterations, mutations summarized at the gene level, and position-specific mutations were evaluated for association with tumor immune infiltration. We observed a strong relationship between copy number loss of a large region of chromosome 9p and decreased lymphocyte estimates in melanoma, pancreatic, and head/neck cancers. Mutations in the oncogenes PIK3CA, FGFR3, and RAS/RAF family members, as well as the tumor suppressor TP53, were linked to changes in immune infiltration, usually in restricted tumor types. Associations of specific WNT/beta-catenin pathway genetic changes with immune state were limited, but we noted a link between 9p loss and the expression of the WNT receptor FZD3, suggesting that there are interactions between 9p alteration and WNT pathways. Finally, two different cell death regulators, CASP8 and DIDO1, were often mutated in head/neck tumors that had higher lymphocyte infiltrates. In summary, our study supports the relevance of tumor genetics to questions of efficacy and resistance in checkpoint blockade therapies. It also highlights the need to assess genome-wide influences during exploration of any specific tumor pathway hypothesized to be relevant to therapeutic response. Some of the observed genetic links to immune state, like 9p loss, may influence response to cancer immune therapies. Others, like mutations in cell death pathways, may help guide combination therapeutic approaches.
Conflict of interest statement
Figures

References
-
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nature reviews Immunology. 2015;15: 486–499. doi: 10.1038/nri3862 - DOI - PMC - PubMed
-
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369: 122–133. doi: 10.1056/NEJMoa1302369 - DOI - PMC - PubMed
-
- Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33: 1889–1894. doi: 10.1200/JCO.2014.56.2736 - DOI - PMC - PubMed
-
- Ji R-R, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer immunology, immunotherapy: CII. 2012;61: 1019–1031. doi: 10.1007/s00262-011-1172-6 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

