Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity
- PMID: 28751364
- PMCID: PMC5565439
- DOI: 10.1073/pnas.1704955114
Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity
Abstract
Nuclear hormone receptors (NRs) regulate physiology by sensing lipophilic ligands and adapting cellular transcription appropriately. A growing understanding of the impact of circadian clocks on mammalian transcription has sparked interest in the interregulation of transcriptional programs. Mammalian clocks are based on a transcriptional feedback loop featuring the transcriptional activators circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1), and transcriptional repressors cryptochrome (CRY) and period (PER). CRY1 and CRY2 bind independently of other core clock factors to many genomic sites, which are enriched for NR recognition motifs. Here we report that CRY1/2 serve as corepressors for many NRs, indicating a new facet of circadian control of NR-mediated regulation of metabolism and physiology, and specifically contribute to diurnal modulation of drug metabolism.
Keywords: circadian; corepressor; cryptochrome; nuclear hormone receptor; xenobiotic metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

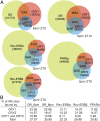
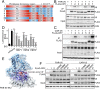


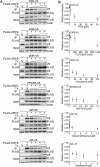
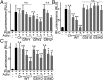
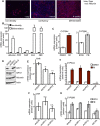
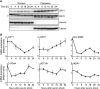

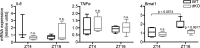
References
-
- Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

