Kinetic analyses of single-stranded break repair by human DNA ligase III isoforms reveal biochemical differences from DNA ligase I
- PMID: 28751376
- PMCID: PMC5612117
- DOI: 10.1074/jbc.M117.804625
Kinetic analyses of single-stranded break repair by human DNA ligase III isoforms reveal biochemical differences from DNA ligase I
Abstract
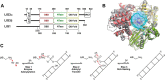
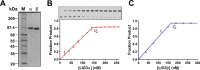
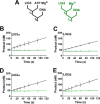
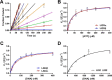
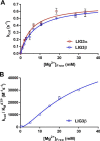
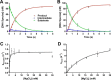
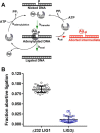
Humans have three genes encoding DNA ligases with conserved structural features and activities, but they also have notable differences. The LIG3 gene encodes a ubiquitous isoform in all tissues (LIG3α) and a germ line-specific splicing isoform (LIG3β) that differs in the C-terminal domain. Both isoforms are found in the nucleus and the mitochondria. Here, we determined the kinetics and thermodynamics of single-stranded break ligation by LIG3α and LIG3β and compared this framework to that of LIG1, the nuclear replicative ligase. The kinetic parameters of the LIG3 isoforms are nearly identical under all tested conditions, indicating that the BRCA1 C terminal (BRCT) domain specific to LIG3α does not alter ligation kinetics. Although LIG3 is only 22% identical to LIG1 across their conserved domains, the two enzymes had very similar maximal ligation rates. Comparison of the rate and equilibrium constants for LIG3 and LIG1 nevertheless revealed important differences. The LIG3 isoforms were seven times more efficient than LIG1 at ligating nicked DNA under optimal conditions, mainly because of their lower Km value for the DNA substrate. This could explain why LIG3 is less prone to abortive ligation than LIG1. Surprisingly, the affinity of LIG3 for Mg2+ was ten times weaker than that of LIG1, suggesting that Mg2+ availability regulates DNA ligation in vivo, because Mg2+ levels are higher in the mitochondria than in the nucleus. The biochemical differences between the LIG3 isoforms and LIG1 identified here will guide the understanding of both unique and overlapping biological roles of these critical enzymes.
Keywords: DNA ligase; DNA repair; DNA replication; abortive ligation; enzyme catalysis; magnesium; pre-steady-state kinetics; steady-state; steady-state kinetics.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
-
- Bentley D., Selfridge J., Millar J. K., Samuel K., Hole N., Ansell J. D., and Melton D. W. (1996) DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 13, 489–491 - PubMed
-
- Bentley D. J., Harrison C., Ketchen A. M., Redhead N. J., Samuel K., Waterfall M., Ansell J. D., and Melton D. W. (2002) DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci. 115, 1551–1561 - PubMed
-
- Barnes D. E., Stamp G., Rosewell I., Denzel A., and Lindahl T. (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8, 1395–1398 - PubMed
-
- Frank K. M., Sekiguchi J. M., Seidl K. J., Swat W., Rathbun G. A., Cheng H. L., Davidson L., Kangaloo L., and Alt F. W. (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396, 173–177 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

