Genome-Wide SNP Discovery and Analysis of Genetic Diversity in Farmed Sika Deer (Cervus nippon) in Northeast China Using Double-Digest Restriction Site-Associated DNA Sequencing
- PMID: 28751500
- PMCID: PMC5592941
- DOI: 10.1534/g3.117.300082
Genome-Wide SNP Discovery and Analysis of Genetic Diversity in Farmed Sika Deer (Cervus nippon) in Northeast China Using Double-Digest Restriction Site-Associated DNA Sequencing
Abstract
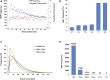
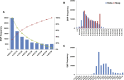
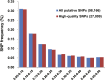
Sika deer are an economically valuable species owing to their use in traditional Chinese medicine, particularly their velvet antlers. Sika deer in northeast China are mostly farmed in enclosure. Therefore, genetic management of farmed sika deer would benefit from detailed knowledge of their genetic diversity. In this study, we generated over 1.45 billion high-quality paired-end reads (288 Gbp) across 42 unrelated individuals using double-digest restriction site-associated DNA sequencing (ddRAD-seq). A total of 96,188 (29.63%) putative biallelic SNP loci were identified with an average sequencing depth of 23×. Based on the analysis, we found that the majority of the loci had a deficit of heterozygotes (FIS >0) and low values of Hobs, which could be due to inbreeding and Wahlund effects. We also developed a collection of high-quality SNP probes that will likely be useful in a variety of applications in genotyping for cervid species in the future.
Keywords: GenPred; Genomic Selection; SNP discovery; STACKS; Shared Data Resources; ddRAD-seq; genetic diversity; sika deer.
Copyright © 2017 Ba et al.
Figures

Similar articles
-
Development of Diagnostic SNP Markers To Monitor Hybridization Between Sika Deer (Cervus nippon) and Wapiti (Cervus elaphus).G3 (Bethesda). 2018 Jul 2;8(7):2173-2179. doi: 10.1534/g3.118.200417. G3 (Bethesda). 2018. PMID: 29789312 Free PMC article.
-
Development and validation of a 1 K sika deer (Cervus nippon) SNP Chip.BMC Genom Data. 2021 Sep 17;22(1):35. doi: 10.1186/s12863-021-00994-z. BMC Genom Data. 2021. PMID: 34535071 Free PMC article.
-
Isolation of novel microsatellite markers and their application for genetic diversity and parentage analyses in sika deer.Gene. 2018 Feb 15;643:68-73. doi: 10.1016/j.gene.2017.12.007. Epub 2017 Dec 6. Gene. 2018. PMID: 29223356
-
Progress on deer genome research.Yi Chuan. 2021 Apr 20;43(4):308-322. doi: 10.16288/j.yczz.20-362. Yi Chuan. 2021. PMID: 33972206 Review.
-
Genetic selection in farmed deer.Vet Rec. 1991 Feb 2;128(5):100-2. doi: 10.1136/vr.128.5.100. Vet Rec. 1991. PMID: 2024412 Review.
Cited by
-
Genome-wide study on genetic diversity and phylogeny of five species in the genus Cervus.BMC Genomics. 2019 May 17;20(1):384. doi: 10.1186/s12864-019-5785-z. BMC Genomics. 2019. PMID: 31101010 Free PMC article.
-
Genome-wide scan for SNPs and selective sweeps reveals candidate genes and QTLs for milk production and reproduction traits in Indian Kankrej cattle.3 Biotech. 2025 Apr;15(4):90. doi: 10.1007/s13205-025-04263-z. Epub 2025 Mar 14. 3 Biotech. 2025. PMID: 40092452
-
SNP discovery of Korean short day onion inbred lines using double digest restriction site-associated DNA sequencing.PLoS One. 2018 Aug 7;13(8):e0201229. doi: 10.1371/journal.pone.0201229. eCollection 2018. PLoS One. 2018. PMID: 30086138 Free PMC article.
-
Genetic features of Sri Lankan elephant, Elephas maximus maximus Linnaeus revealed by high throughput sequencing of mitogenome and ddRAD-seq.PLoS One. 2023 Jun 13;18(6):e0285572. doi: 10.1371/journal.pone.0285572. eCollection 2023. PLoS One. 2023. PMID: 37310948 Free PMC article.
-
Integrated mRNA and miRNA Analysis Reveals Layer-Specific Mechanisms of Antler Yield Variation in Sika Deer.Animals (Basel). 2025 Jul 4;15(13):1964. doi: 10.3390/ani15131964. Animals (Basel). 2025. PMID: 40646863 Free PMC article.
References
-
- Ba H., Yang F., Xing X., Li C., 2015. Classification and phylogeny of sika deer (Cervus nippon) subspecies based on the mitochondrial control region DNA sequence using an extended sample set. Mitochondrial DNA 26: 373–379. - PubMed
-
- Ba H., Wu L., Liu Z., Li C., 2016. An examination of the origin and evolution of additional tandem repeats in the mitochondrial DNA control region of Japanese sika deer (Cervus Nippon). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27: 276–281. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
