The Douglas-Fir Genome Sequence Reveals Specialization of the Photosynthetic Apparatus in Pinaceae
- PMID: 28751502
- PMCID: PMC5592940
- DOI: 10.1534/g3.117.300078
The Douglas-Fir Genome Sequence Reveals Specialization of the Photosynthetic Apparatus in Pinaceae
Abstract
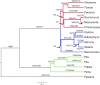
A reference genome sequence for Pseudotsuga menziesii var. menziesii (Mirb.) Franco (Coastal Douglas-fir) is reported, thus providing a reference sequence for a third genus of the family Pinaceae. The contiguity and quality of the genome assembly far exceeds that of other conifer reference genome sequences (contig N50 = 44,136 bp and scaffold N50 = 340,704 bp). Incremental improvements in sequencing and assembly technologies are in part responsible for the higher quality reference genome, but it may also be due to a slightly lower exact repeat content in Douglas-fir vs. pine and spruce. Comparative genome annotation with angiosperm species reveals gene-family expansion and contraction in Douglas-fir and other conifers which may account for some of the major morphological and physiological differences between the two major plant groups. Notable differences in the size of the NDH-complex gene family and genes underlying the functional basis of shade tolerance/intolerance were observed. This reference genome sequence not only provides an important resource for Douglas-fir breeders and geneticists but also sheds additional light on the evolutionary processes that have led to the divergence of modern angiosperms from the more ancient gymnosperms.
Keywords: annotation; conifer; genome assembly; gymnosperm; mega-genome; shade tolerance.
Copyright © 2017 Neale et al.
Figures


Similar articles
-
A long-read and short-read transcriptomics approach provides the first high-quality reference transcriptome and genome annotation for Pseudotsuga menziesii (Douglas-fir).G3 (Bethesda). 2023 Feb 9;13(2):jkac304. doi: 10.1093/g3journal/jkac304. G3 (Bethesda). 2023. PMID: 36454025 Free PMC article.
-
Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies.Genome Biol Evol. 2010;2:504-17. doi: 10.1093/gbe/evq036. Epub 2010 Jul 2. Genome Biol Evol. 2010. PMID: 20651328 Free PMC article.
-
A mutation hotspot in the chloroplast genome of a conifer (Douglas-fir: Pseudotsuga) is caused by variability in the number of direct repeats derived from a partially duplicated tRNA gene.Curr Genet. 1995 May;27(6):572-9. doi: 10.1007/BF00314450. Curr Genet. 1995. PMID: 7553944
-
Karyotype evolution in the Pinaceae: implication with molecular phylogeny.Genome. 2012 Nov;55(11):735-53. doi: 10.1139/g2012-061. Epub 2012 Oct 25. Genome. 2012. PMID: 23199570 Review.
-
Towards decoding the conifer giga-genome.Plant Mol Biol. 2012 Dec;80(6):555-69. doi: 10.1007/s11103-012-9961-7. Epub 2012 Sep 9. Plant Mol Biol. 2012. PMID: 22960864 Review.
Cited by
-
An Axiom SNP genotyping array for Douglas-fir.BMC Genomics. 2020 Jan 3;21(1):9. doi: 10.1186/s12864-019-6383-9. BMC Genomics. 2020. PMID: 31900111 Free PMC article.
-
Genetic diversity and structure of the 4th cycle breeding population of Chinese fir (Cunninghamia lanceolata (lamb.) hook).Front Plant Sci. 2023 Jan 27;14:1106615. doi: 10.3389/fpls.2023.1106615. eCollection 2023. Front Plant Sci. 2023. PMID: 36778690 Free PMC article.
-
Utilization of Tissue Ploidy Level Variation in de Novo Transcriptome Assembly of Pinus sylvestris.G3 (Bethesda). 2019 Oct 7;9(10):3409-3421. doi: 10.1534/g3.119.400357. G3 (Bethesda). 2019. PMID: 31427456 Free PMC article.
-
A revised view on the evolution of glutamine synthetase isoenzymes in plants.Plant J. 2022 May;110(4):946-960. doi: 10.1111/tpj.15712. Epub 2022 Mar 9. Plant J. 2022. PMID: 35199893 Free PMC article.
-
Water-related innovations in land plants evolved by different patterns of gene cooption and novelty.New Phytol. 2022 Jul;235(2):732-742. doi: 10.1111/nph.17981. Epub 2022 Feb 8. New Phytol. 2022. PMID: 35048381 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
