High resolution temporal transcriptomics of mouse embryoid body development reveals complex expression dynamics of coding and noncoding loci
- PMID: 28751729
- PMCID: PMC5532269
- DOI: 10.1038/s41598-017-06110-5
High resolution temporal transcriptomics of mouse embryoid body development reveals complex expression dynamics of coding and noncoding loci
Abstract
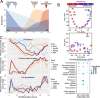
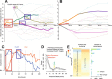
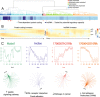
Cellular responses to stimuli are rapid and continuous and yet the vast majority of investigations of transcriptional responses during developmental transitions typically use long interval time courses; limiting the available interpretive power. Moreover, such experiments typically focus on protein-coding transcripts, ignoring the important impact of long noncoding RNAs. We therefore evaluated coding and noncoding expression dynamics at unprecedented temporal resolution (6-hourly) in differentiating mouse embryonic stem cells and report new insight into molecular processes and genome organization. We present a highly resolved differentiation cascade that exhibits coding and noncoding transcriptional alterations, transcription factor network interactions and alternative splicing events, little of which can be resolved by long-interval developmental time-courses. We describe novel short lived and cycling patterns of gene expression and dissect temporally ordered gene expression changes in response to transcription factors. We elucidate patterns in gene co-expression across the genome, describe asynchronous transcription at bidirectional promoters and functionally annotate known and novel regulatory lncRNAs. These findings highlight the complex and dynamic molecular events underlying mammalian differentiation that can only be observed though a temporally resolved time course.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

