Simultaneous Methylation-Level Assessment of Hundreds of CpG Sites by Targeted Bisulfite PCR Sequencing (TBPseq)
- PMID: 28751909
- PMCID: PMC5507944
- DOI: 10.3389/fgene.2017.00097
Simultaneous Methylation-Level Assessment of Hundreds of CpG Sites by Targeted Bisulfite PCR Sequencing (TBPseq)
Abstract
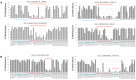
Methylated-DNA sequencing technologies are producing vast amounts of methylome data from cancer samples, from which cancer-associated differentially methylated CpG sites (cDMCs) are continuously identified and filed. The inclusion of as many cDMCs as possible helps improve the accuracy of cancer diagnosis and sometimes identify cancer subtypes. However, the lack of an established method for the analysis of 100s of cDMCs practically impedes their robust use in clinical medicine. Here, we tested the availability of targeted bisulfite-PCR-sequencing (TBPseq) technology for the assessment of methylation levels of a myriad of CpGs scattered over the genome. In randomly selected 46 cancer cell lines, multiplexed PCR yielded a variety of amplicons harboring 246 CpGs residing at promoters of 97 cancer-associated genes, all of which were sequenced in the same flow cell. Clustering analysis of the TBPseq-assessed methylation levels of target CpGs showed that the lung and liver cancer cell lines correlated relatively strongly with each other while they weakly correlated with colon cancer cells. CpGs at the LIFR gene promoter, which are known to be hypermethylated in colon cancers, indeed were heavily methylated in the tested colon cancer cells. Moreover, the LIFR promoter hypermethylation was found in colon cancer cells only, but not in biliary tract, liver, lung, and stomach cancers cell lines. A meta-analysis with public cancer methylome data verified the colon cancer specificity of LIFR promoter methylation. These results demonstrate that our TBPseq-based methylation assessment could be considered an effective, accurate, and competitive method to simultaneously examine a large number of target cDMCs and patient samples.
Keywords: DNA methylation; LIFR; cancer; diagnosis; sequencing; targeted NGS.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

