The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease
- PMID: 28752201
- PMCID: PMC5756263
- DOI: 10.1007/s00018-017-2598-6
The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease
Abstract
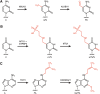
Mitochondrial protein synthesis is essential for the production of components of the oxidative phosphorylation system. RNA modifications in the mammalian mitochondrial translation apparatus play key roles in facilitating mitochondrial gene expression as they enable decoding of the non-conventional genetic code by a minimal set of tRNAs, and efficient and accurate protein synthesis by the mitoribosome. Intriguingly, recent transcriptome-wide analyses have also revealed modifications in mitochondrial mRNAs, suggesting that the concept of dynamic regulation of gene expression by the modified RNAs (the "epitranscriptome") extends to mitochondria. Furthermore, it has emerged that defects in RNA modification, arising from either mt-DNA mutations or mutations in nuclear-encoded mitochondrial modification enzymes, underlie multiple mitochondrial diseases. Concomitant advances in the identification of the mitochondrial RNA modification machinery and recent structural views of the mitochondrial translation apparatus now allow the molecular basis of such mitochondrial diseases to be understood on a mechanistic level.
Keywords: Epitranscriptome; Mitochondria; Mitochondrial disease; Protein synthesis; RNA modification; Ribosome; Translation; tRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C (2012) Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13:566–578 - PubMed
-
- Nasrallah CM, Horvath TL (2014) Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol 10:650–658 - PubMed
-
- Lang BF, Gray MW, Burger G (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet 33:351–397 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

