The first phlebo-like virus infecting plants: a case study on the adaptation of negative-stranded RNA viruses to new hosts
- PMID: 28752569
- PMCID: PMC6637980
- DOI: 10.1111/mpp.12587
The first phlebo-like virus infecting plants: a case study on the adaptation of negative-stranded RNA viruses to new hosts
Abstract
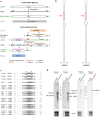
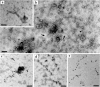
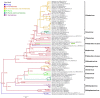
A novel negative-stranded (ns) RNA virus associated with a severe citrus disease reported more than 80 years ago has been identified. Transmission electron microscopy showed that this novel virus, tentatively named citrus concave gum-associated virus, is flexuous and non-enveloped. Notwithstanding, its two genomic RNAs share structural features with members of the genus Phlebovirus, which are enveloped arthropod-transmitted viruses infecting mammals, and with a group of still unclassified phlebo-like viruses mainly infecting arthropods. CCGaV genomic RNAs code for an RNA-dependent RNA polymerase, a nucleocapsid protein and a putative movement protein showing structural and phylogenetic relationships with phlebo-like viruses, phleboviruses and the unrelated ophioviruses, respectively, thus providing intriguing evidence of a modular genome evolution. Phylogenetic reconstructions identified an invertebrate-restricted virus as the most likely ancestor of this virus, revealing that its adaptation to plants was independent from and possibly predated that of the other nsRNA plant viruses. These data are consistent with an evolutionary scenario in which trans-kingdom adaptation occurred several times during the history of nsRNA viruses and followed different evolutionary pathways, in which genomic RNA segments were gained or lost. The need to create a new genus for this bipartite nsRNA virus and the impact of the rapid and specific detection methods developed here on citrus sanitation and certification are also discussed.
Keywords: citrus disease; concave gum; nsRNA virus; phylogeny; virus adaptation.
© 2017 BSPP AND JOHN WILEY & SONS LTD.
Figures



References
-
- Abascal, F. , Zardoya, R. and Posada, D. (2005) ProtTest: selection of best‐fit models of protein evolution. Bioinformatics, 21, 2104–2105. - PubMed
-
- Alioto, D. , Gangemi, M. , Deaglio, S. , Sposato, P. , Noris, E. , Luisoni, E. and Milne, R.G. (1999) Improved detection of citrus psorosis virus using polyclonal and monoclonal antibodies. Plant Pathol. 48, 735–741.
-
- Bekal, S. , Domier, L.L. , Niblack, T.L. and Lambert, K.N. (2011) Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J. Gen. Virol. 92, 1870–1879. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

