New findings on phosphodiesterases, MoPdeH and MoPdeL, in Magnaporthe oryzae revealed by structural analysis
- PMID: 28752677
- PMCID: PMC6638029
- DOI: 10.1111/mpp.12586
New findings on phosphodiesterases, MoPdeH and MoPdeL, in Magnaporthe oryzae revealed by structural analysis
Abstract
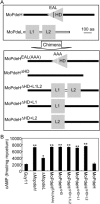
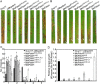
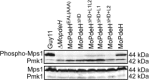
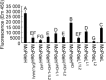
The cyclic adenosine monophosphate (cAMP) signalling pathway mediates signal communication and sensing during infection-related morphogenesis in eukaryotes. Many studies have implicated cAMP as a critical mediator of appressorium development in the rice blast fungus, Magnaporthe oryzae. The cAMP phosphodiesterases, MoPdeH and MoPdeL, as key regulators of intracellular cAMP levels, play pleiotropic roles in cell wall integrity, cellular morphology, appressorium formation and infectious growth in M. oryzae. Here, we analysed the roles of domains of MoPdeH and MoPdeL separately or in chimeras. The results indicated that the HD and EAL domains of MoPdeH are indispensable for its phosphodiesterase activity and function. Replacement of the MoPdeH HD domain with the L1 and L2 domains of MoPdeL, either singly or together, resulted in decreased cAMP hydrolysis activity of MoPdeH. All of the transformants exhibited phenotypes similar to that of the ΔMopdeH mutant, but also revealed that EAL and L1 play additional roles in conidiation, and that L1 is involved in infectious growth. We further found that the intracellular cAMP level is important for surface signal recognition and hyphal autolysis. The intracellular cAMP level negatively regulates Mps1-MAPK and positively regulates Pmk1-MAPK in the rice blast fungus. Our results provide new information to better understand the cAMP signalling pathway in the development, differentiation and plant infection of the fungus.
Keywords: MoPdeH/L; appressorium formation; cAMP level; pathogenicity; phosphodiesterase activity.
© 2017 BSPP AND JOHN WILEY & SONS LTD.
Figures







References
-
- Arnold, K. , Bordoli, L. , Kopp, J. and Schwede, T. (2006) The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics, 22, 195–201. - PubMed
-
- Bencina, M. , Panneman, H. , Ruijter, G.J. , Legisa, M. and Visser, J. (1997) Characterization and overexpression of the Aspergillus niger gene encoding the cAMP‐dependent protein kinase catalytic subunit. Microbiology, 143, 1211–1220. - PubMed
-
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , Kiefer, F. , Gallo Cassarino, T. , Bertoni, M. , Bordoli, L. and Schwede, T. (2014) SWISS‐MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. - PMC - PubMed
-
- Bobrov, A.G. , Kirillina, O. and Perry, R.D. (2005) The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis . FEMS Microbiol. Lett. 247, 123–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

