Systematic genetic dissection of chitin degradation and uptake in Vibrio cholerae
- PMID: 28752963
- PMCID: PMC5647239
- DOI: 10.1111/1462-2920.13866
Systematic genetic dissection of chitin degradation and uptake in Vibrio cholerae
Abstract
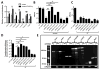
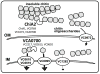
Vibrio cholerae is a natural resident of the aquatic environment, where a common nutrient is the chitinous exoskeletons of microscopic crustaceans. Chitin utilization requires chitinases, which degrade this insoluble polymer into soluble chitin oligosaccharides. These oligosaccharides also serve as an inducing cue for natural transformation in Vibrio species. There are 7 predicted endochitinase-like genes in the V. cholerae genome. Here, we systematically dissect the contribution of each gene to growth on chitin as well as induction of natural transformation. Specifically, we created a strain that lacks all 7 putative chitinases and from this strain, generated a panel of strains where each expresses a single chitinase. We also generated expression plasmids to ectopically express all 7 chitinases in our chitinase deficient strain. Through this analysis, we found that low levels of chitinase activity are sufficient for natural transformation, while growth on insoluble chitin as a sole carbon source requires more robust and concerted chitinase activity. We also assessed the role that the three uptake systems for the chitin degradation products GlcNAc, (GlcNAc)2 and (GlcN)2 , play in chitin utilization and competence induction. Cumulatively, this study provides mechanistic details for how this pathogen utilizes chitin to thrive and evolve in its environmental reservoir.
© 2017 Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures
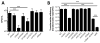
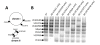
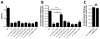
References
-
- Bassler BL, Yu C, Lee YC, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. - PubMed
-
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. - PubMed
-
- Dalia AB. RpoS is required for natural transformation of Vibrio cholerae through regulation of chitinases. Environ Microbiol. 2016;18:3758–3767. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

