How Ligands Illuminate GPCR Molecular Pharmacology
- PMID: 28753422
- PMCID: PMC5560499
- DOI: 10.1016/j.cell.2017.07.009
How Ligands Illuminate GPCR Molecular Pharmacology
Abstract
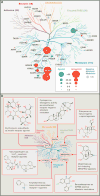
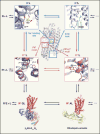
G protein-coupled receptors (GPCRs), which are modulated by a variety of endogenous and synthetic ligands, represent the largest family of druggable targets in the human genome. Recent structural and molecular studies have both transformed and expanded classical concepts of receptor pharmacology and have begun to illuminate the distinct mechanisms by which structurally, chemically, and functionally diverse ligands modulate GPCR function. These molecular insights into ligand engagement and action have enabled new computational methods and accelerated the discovery of novel ligands and tool compounds, especially for understudied and orphan GPCRs. These advances promise to streamline the development of GPCR-targeted medications.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. - PubMed
-
- Aitken M, Berndt ER, Cutler DM. Prescription drug spending trends in the United States: looking beyond the turning point. Health Aff (Millwood) 2009;28:w151–160. - PubMed
-
- Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011a;51:117–144. - PubMed
-
- Allen JA, Yost JM, Setola V, Chen X, Sassano MF, Chen M, Peterson S, Yadav PN, Huang XP, Feng B, Jensen NH, Che X, Bai X, Frye SV, Wetsel WC, Caron MG, Javitch JA, Roth BL, Jin J. Discovery of beta-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc Natl Acad Sci U S A. 2011b;108:18488–18493. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

