Modulation of kanamycin B and kanamycin A biosynthesis in Streptomyces kanamyceticus via metabolic engineering
- PMID: 28753625
- PMCID: PMC5533434
- DOI: 10.1371/journal.pone.0181971
Modulation of kanamycin B and kanamycin A biosynthesis in Streptomyces kanamyceticus via metabolic engineering
Abstract
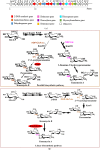
Both kanamycin A and kanamycin B, antibiotic components produced by Streptomyces kanamyceticus, have medical value. Two different pathways for kanamycin biosynthesis have been reported by two research groups. In this study, to obtain an optimal kanamycin A-producing strain and a kanamycin B-high-yield strain, we first examined the native kanamycin biosynthetic pathway in vivo. Based on the proposed parallel biosynthetic pathway, kanN disruption should lead to kanamycin A accumulation; however, the kanN-disruption strain produced neither kanamycin A nor kanamycin B. We then tested the function of kanJ and kanK. The main metabolite of the kanJ-disruption strain was identified as kanamycin B. These results clarified that kanamycin biosynthesis does not proceed through the parallel pathway and that synthesis of kanamycin A from kanamycin B is catalyzed by KanJ and KanK in S. kanamyceticus. As expected, the kanamycin B yield of the kanJ-disruption strain was 3268±255 μg/mL, 12-fold higher than that of the original strain. To improve the purity of kanamycin A and reduce the yield of kanamycin B in the fermentation broth, four different kanJ- and kanK-overexpressing strains were constructed through either homologous recombination or site-specific integration. The overexpressing strain containing three copies of kanJ and kanK in its genome exhibited the lowest kanamycin B yield (128±20 μg/mL), which was 54% lower than that of the original strain. Our experimental results demonstrate that kanamycin A is derived from KanJ-and-KanK-catalyzed conversion of kanamycin B in S. kanamyceticus. Moreover, based on the clarified biosynthetic pathway, we obtained a kanamycin B-high-yield strain and an optimized kanamycin A-producing strain with minimal byproduct.
Conflict of interest statement
Figures





Similar articles
-
Establishment of a highly efficient conjugation protocol for Streptomyces kanamyceticus ATCC12853.Microbiologyopen. 2019 Jun;8(6):e00747. doi: 10.1002/mbo3.747. Epub 2018 Nov 17. Microbiologyopen. 2019. PMID: 30449069 Free PMC article.
-
Construction of kanamycin B overproducing strain by genetic engineering of Streptomyces tenebrarius.Appl Microbiol Biotechnol. 2011 Feb;89(3):723-31. doi: 10.1007/s00253-010-2908-5. Epub 2010 Oct 9. Appl Microbiol Biotechnol. 2011. PMID: 20936279
-
The last step of kanamycin biosynthesis: unique deamination reaction catalyzed by the α-ketoglutarate-dependent nonheme iron dioxygenase KanJ and the NADPH-dependent reductase KanK.Angew Chem Int Ed Engl. 2012 Apr 2;51(14):3428-31. doi: 10.1002/anie.201108122. Epub 2012 Feb 28. Angew Chem Int Ed Engl. 2012. PMID: 22374809
-
Streptomyces species: Ideal chassis for natural product discovery and overproduction.Metab Eng. 2018 Nov;50:74-84. doi: 10.1016/j.ymben.2018.05.015. Epub 2018 May 28. Metab Eng. 2018. PMID: 29852270 Review.
-
Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement.J Antibiot (Tokyo). 2013 Jul;66(7):411-20. doi: 10.1038/ja.2013.26. Epub 2013 Apr 24. J Antibiot (Tokyo). 2013. PMID: 23612724 Review.
Cited by
-
A Review of the Microbial Production of Bioactive Natural Products and Biologics.Front Microbiol. 2019 Jun 20;10:1404. doi: 10.3389/fmicb.2019.01404. eCollection 2019. Front Microbiol. 2019. PMID: 31281299 Free PMC article. Review.
-
Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.Nat Chem Biol. 2019 Mar;15(3):295-303. doi: 10.1038/s41589-018-0203-4. Epub 2019 Jan 14. Nat Chem Biol. 2019. PMID: 30643280 Free PMC article.
-
Establishment of a highly efficient conjugation protocol for Streptomyces kanamyceticus ATCC12853.Microbiologyopen. 2019 Jun;8(6):e00747. doi: 10.1002/mbo3.747. Epub 2018 Nov 17. Microbiologyopen. 2019. PMID: 30449069 Free PMC article.
-
Characterization and utilization of methyltransferase for apramycin production in Streptoalloteichus tenebrarius.J Ind Microbiol Biotechnol. 2022 Jul 30;49(4):kuac011. doi: 10.1093/jimb/kuac011. J Ind Microbiol Biotechnol. 2022. PMID: 35536571 Free PMC article.
-
Improved production of clavulanic acid by reverse engineering and overexpression of the regulatory genes in an industrial Streptomyces clavuligerus strain.J Ind Microbiol Biotechnol. 2019 Aug;46(8):1205-1215. doi: 10.1007/s10295-019-02196-0. Epub 2019 Jun 4. J Ind Microbiol Biotechnol. 2019. PMID: 31165280
References
-
- Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54: 287–301. doi: 10.1007/s002530000403 - DOI - PubMed
-
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407: 340–348. doi: 10.1038/35030019 - DOI - PubMed
-
- Kondo S, Hotta K. Semisynthetic aminoglycoside antibiotics: development and enzymatic modifications. J Infect Chemother. 1999;5: 1–9. doi: 10.1007/s101569900000 - DOI - PubMed
-
- Kondo S, Iinuma K, Yamamoto H, Maeda K, Umezawa H. Syntheses of 1-n-(S)-4-amino-2-hydroxybutyryl)-kanamycin B and -3', 4'-dideoxykanamycin B active against kanamycin-resistant bacteria. J Antibiot (Tokyo). 1973;26: 412–415. doi: 10.7164/antibiotics.26.412 - DOI - PubMed
-
- Hsu BH, Orton E, Tang SY, Carlton RA. Application of evaporative light scattering detection to the characterization of combinatorial and parallel synthesis libraries for pharmaceutical drug discovery. J Chromatogr B Biomed Sci Appl. 1999;725: 103–112. doi: 10.1016/S0378-4347(98)00529-5 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

