A novel mechanism for variable phenotypic expressivity in Mendelian diseases uncovered by an AU-rich element (ARE)-creating mutation
- PMID: 28754144
- PMCID: PMC5534118
- DOI: 10.1186/s13059-017-1274-3
A novel mechanism for variable phenotypic expressivity in Mendelian diseases uncovered by an AU-rich element (ARE)-creating mutation
Abstract
Background: Variable expressivity is a well-known phenomenon in which patients with mutations in one gene display varying degrees of clinical severity, potentially displaying only subsets of the clinical manifestations associated with the multisystem disorder linked to the gene. This remains an incompletely understood phenomenon with proposed mechanisms ranging from allele-specific to stochastic.
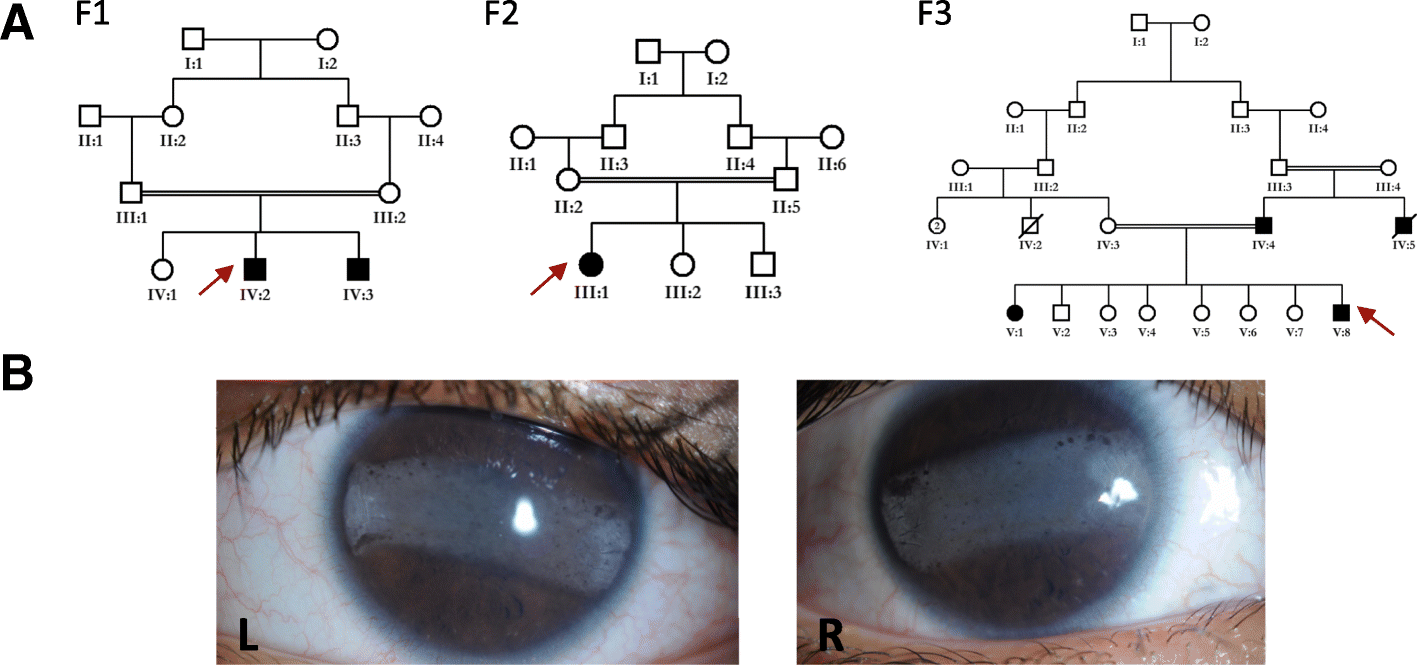
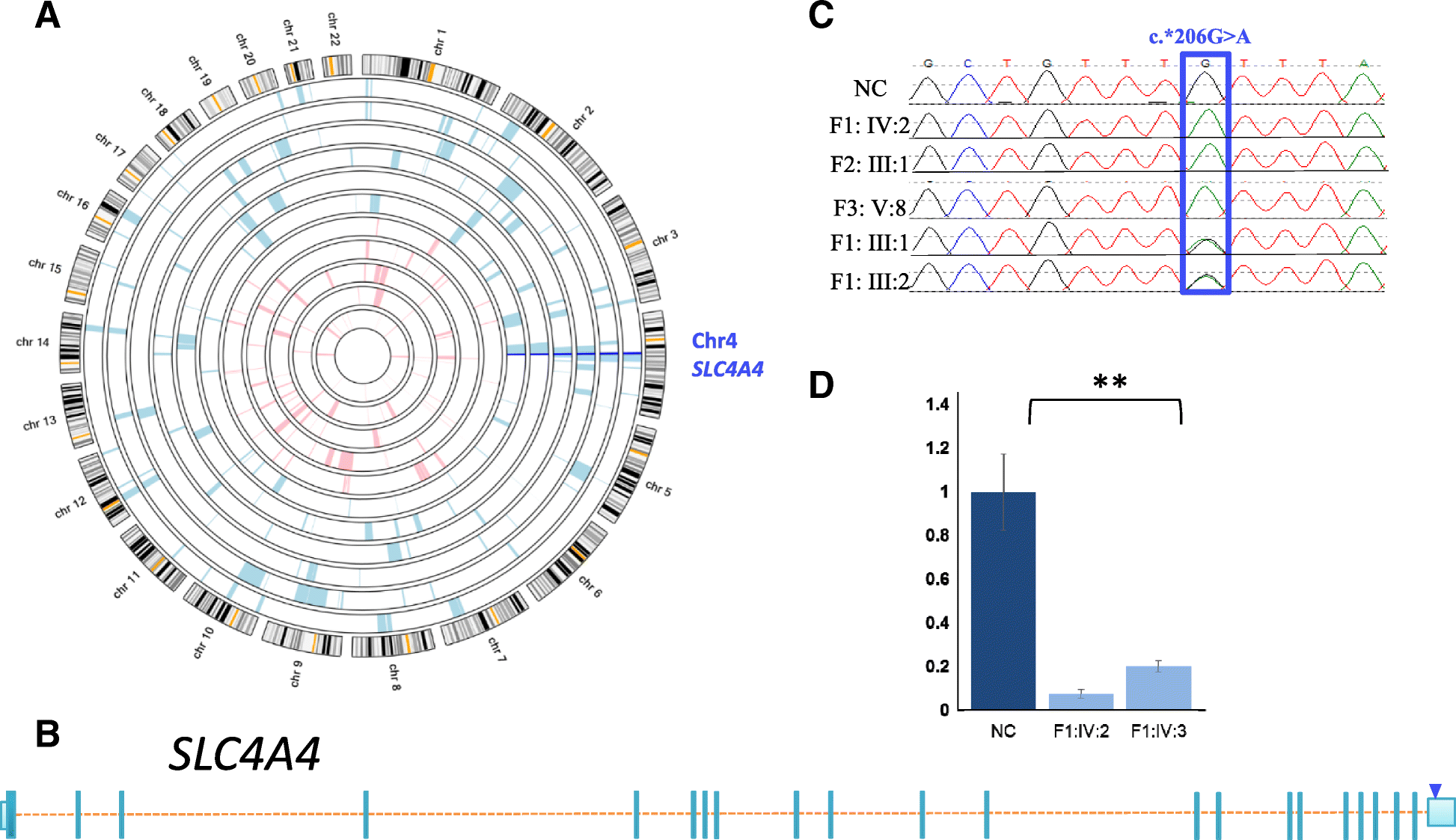
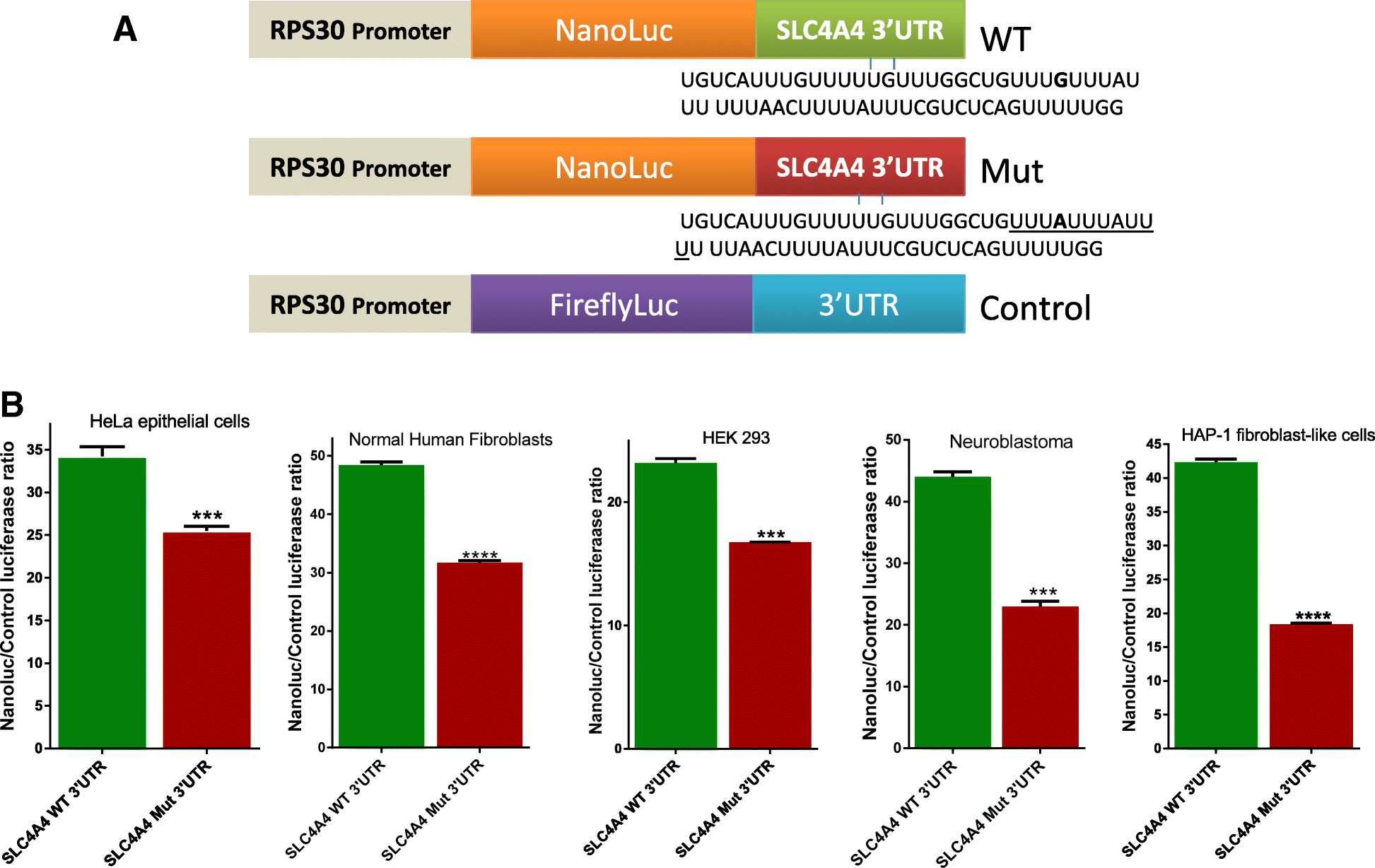
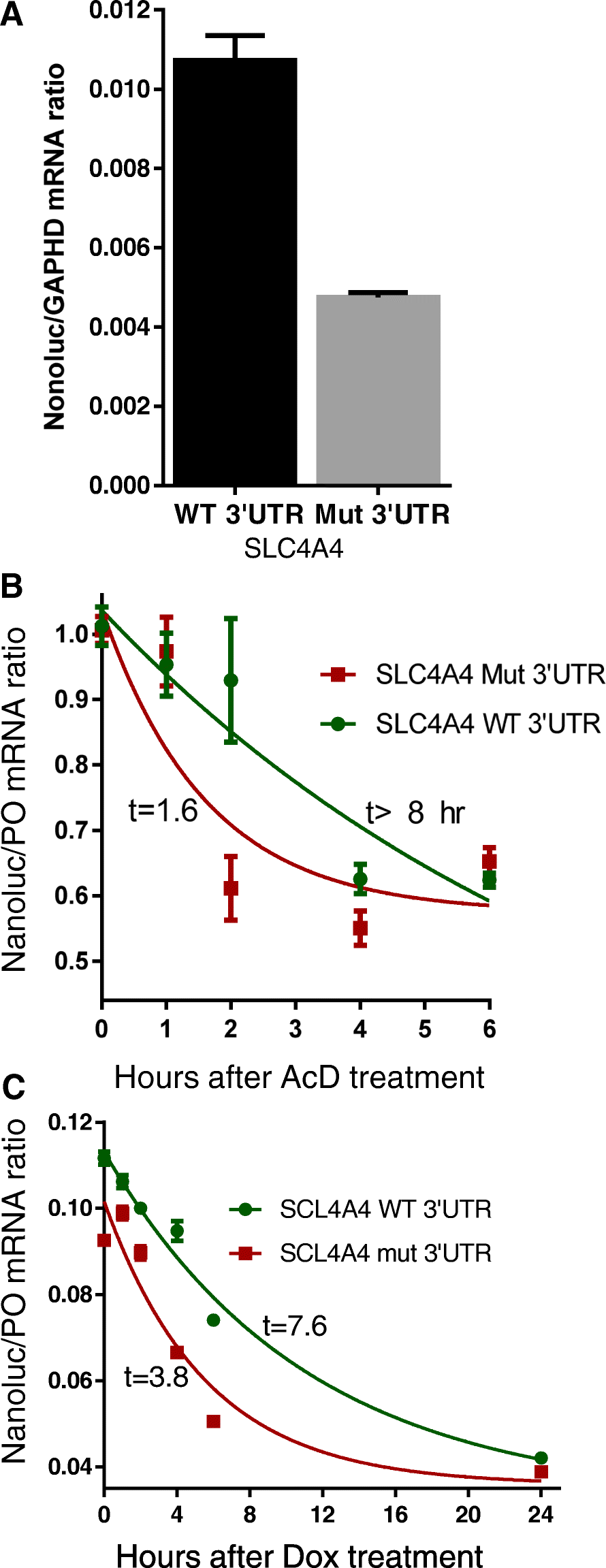
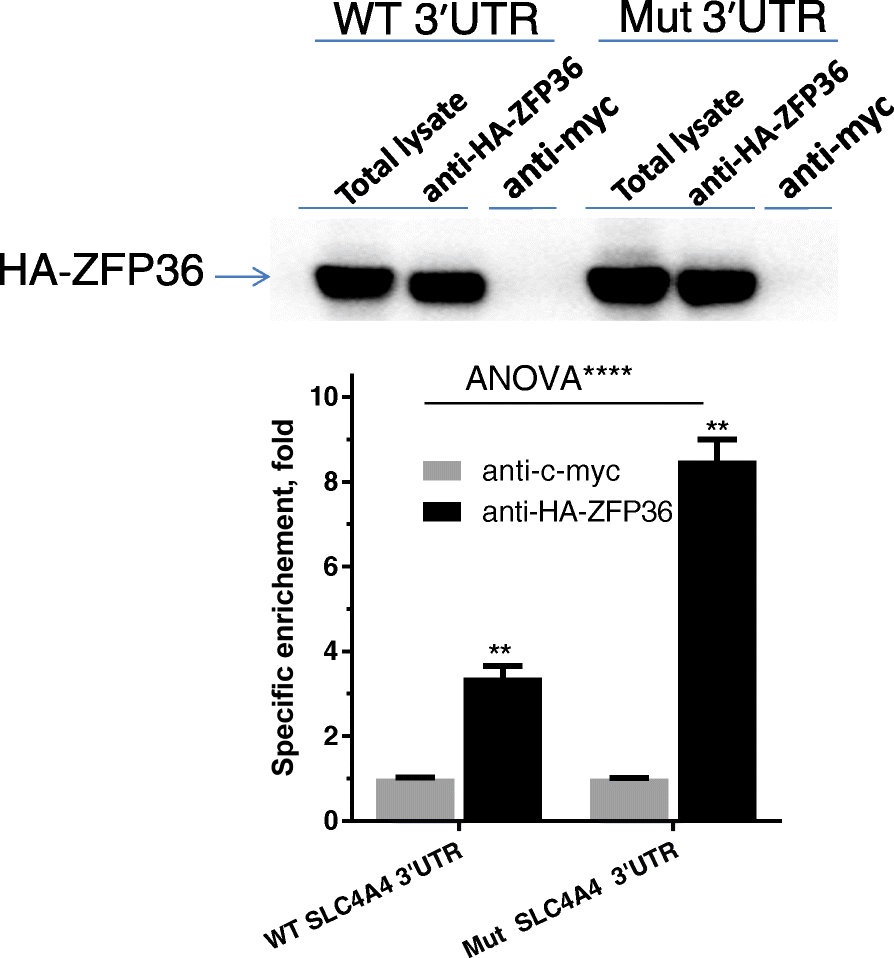
Results: We report three consanguineous families in which an isolated ocular phenotype is linked to a novel 3' UTR mutation in SLC4A4, a gene known to be mutated in a syndromic form of intellectual disability with renal and ocular involvement. Although SLC4A4 is normally devoid of AU-rich elements (AREs), a 3' UTR motif that mediates post-transcriptional control of a subset of genes, the mutation we describe creates a functional ARE. We observe a marked reduction in the transcript level of SLC4A4 in patient cells. Experimental confirmation of the ARE-creating mutation is shown using a post-transcriptional reporter system that reveals consistent reduction in the mRNA-half life and reporter activity. Moreover, the neo-ARE binds and responds to the zinc finger protein ZFP36/TTP, an ARE-mRNA decay-promoting protein.
Conclusions: This novel mutational mechanism for a Mendelian disease expands the potential mechanisms that underlie variable phenotypic expressivity in humans to also include 3' UTR mutations with tissue-specific pathology.
Keywords: 3′UTR; AU-rich elements; Cornea; Tissue-specific; Variable expressivity.
Conflict of interest statement
Ethics approval and consent to participate
This study was IRB-approved (KFSRHC RAC# 2070023). All subjects have given written informed consent and all experimental methods comply with the Helsinki Declaration.
Competing interests
The authors declare that they have no competing interests.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

