The Hsp70 interdomain linker is a dynamic switch that enables allosteric communication between two structured domains
- PMID: 28754691
- PMCID: PMC5592658
- DOI: 10.1074/jbc.M117.789313
The Hsp70 interdomain linker is a dynamic switch that enables allosteric communication between two structured domains
Abstract
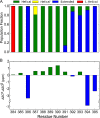
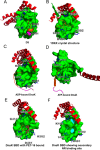
Hsp70 molecular chaperones play key roles in cellular protein homeostasis by binding to exposed hydrophobic regions of incompletely folded or aggregated proteins. This crucial Hsp70 function relies on allosteric communication between two well-structured domains: an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD), which are tethered by an interdomain linker. ATP or ADP binding to the NBD alters the substrate-binding affinity of the SBD, triggering functionally essential cycles of substrate binding and release. The interdomain linker is a well-structured participant in the interdomain interface in ATP-bound Hsp70s. By contrast, in the ADP-bound state, exemplified by the Escherichia coli Hsp70 DnaK, the interdomain linker is flexible. Hsp70 interdomain linker sequences are highly conserved; moreover, mutations in this region compromise interdomain allostery. To better understand the role of this region in Hsp70 allostery, we used molecular dynamics simulations to explore the conformational landscape of the interdomain linker in ADP-bound DnaK and supported our simulations by strategic experimental data. We found that while the interdomain linker samples many conformations, it behaves as three relatively ordered segments connected by hinges. As a consequence, the distances and orientations between the NBD and SBD are limited. Additionally, the C-terminal region of the linker forms previously unreported, transient interactions with the SBD, and the predominant linker-docking site is available in only one allosteric state, that with high affinity for substrate. This preferential binding implicates the interdomain linker as a dynamic allosteric switch. The linker-binding site on the SBD is a potential target for small molecule modulators of the Hsp70 allosteric cycle.
Keywords: 70-kilodalton heat shock protein (Hsp70); DnaK; Hsp70; allosteric regulation; allostery; chaperone DnaK (DnaK); conformational simulation; molecular chaperone; molecular dynamics.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Gierasch L. M. (2016) Hsp70 molecular machines: versatile modular nanomachines that mediate multiple biological functions. In Structure and Action of Molecular Chaperones (Gierasch L. M., Horwich A. L., Slingsby C., Wickner S., and Agard D., eds) pp. 1–48, World Scientific, Singapore
-
- Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

