Coronafacoyl Phytotoxin Biosynthesis and Evolution in the Common Scab Pathogen Streptomyces scabiei
- PMID: 28754703
- PMCID: PMC5601335
- DOI: 10.1128/AEM.01169-17
Coronafacoyl Phytotoxin Biosynthesis and Evolution in the Common Scab Pathogen Streptomyces scabiei
Abstract
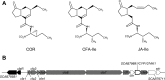
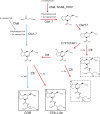
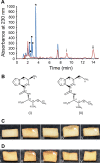
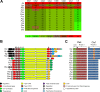
Coronafacoyl phytotoxins are an important family of plant toxins that are produced by several different phytopathogenic bacteria, including the gammaproteobacterium Pseudomonas syringae and the actinobacterium Streptomyces scabiei (formerly Streptomyces scabies). The phytotoxins consist of coronafacic acid (CFA) linked via an amide bond to different amino acids or amino acid derivatives. Previous work suggested that S. scabiei and P. syringae use distinct biosynthetic pathways for producing CFA, which is subsequently linked to its amino acid partner to form the complete phytotoxin. Here, we provide further evidence that the S. scabiei CFA biosynthetic pathway is novel by characterizing the role of CYP107AK1, a predicted cytochrome P450 that has no homologue in P. syringae Deletion of the CYP107AK1 gene abolished production of coronafacoyl-isoleucine (CFA-Ile), the primary coronafacoyl phytotoxin produced by S. scabiei Structural elucidation of accumulated biosynthetic intermediates in the ΔCYP107AK1 mutant indicated that CYP107AK1 is required for introducing the oxygen atom that ultimately forms the carbonyl group in the CFA backbone. The CYP107AK1 gene along with two additional genes involved in CFA-Ile biosynthesis in S. scabiei were found to be associated with putative CFA biosynthetic genes in other actinobacteria but not in other organisms. Analysis of the overall genetic content and organization of known and putative CFA biosynthetic gene clusters, together with phylogenetic analysis of the core biosynthetic genes, indicates that horizontal gene transfer has played an important role in the dissemination of the gene cluster and that rearrangement, insertion, and/or deletion events have likely contributed to the divergent biosynthetic evolution of coronafacoyl phytotoxins in bacteria.IMPORTANCE The ability of plants to defend themselves against invading pathogens relies on complex signaling pathways that are controlled by key phytohormones such as jasmonic acid (JA). Some phytopathogenic bacteria have evolved the ability to manipulate JA signaling in order to overcome host defenses by producing coronatine (COR), which functions as a potent JA mimic. COR and COR-like molecules, collectively referred to as coronafacoyl phytotoxins, are produced by several different plant-pathogenic bacteria, and this study provides supporting evidence that different biosynthetic pathways are utilized by different bacteria for production of these phytotoxins. In addition, our study provides a greater understanding of how coronafacoyl phytotoxin biosynthesis may have evolved in phylogenetically distinct bacteria, and we demonstrate that production of these compounds may be more widespread than previously recognized and that their role for the producing organism may not be limited to host-pathogen interactions.
Keywords: Pseudomonas syringae; Streptomyces scabiei; biosynthesis; coronafacoyl phytotoxin; coronatine; cytochrome P450; gene cluster; jasmonic acid; plant pathogens; signaling.
Copyright © 2017 American Society for Microbiology.
Figures
Similar articles
-
Production of the Streptomyces scabies coronafacoyl phytotoxins involves a novel biosynthetic pathway with an F420 -dependent oxidoreductase and a short-chain dehydrogenase/reductase.Mol Microbiol. 2016 Jul;101(1):122-35. doi: 10.1111/mmi.13378. Epub 2016 Apr 20. Mol Microbiol. 2016. PMID: 26991928
-
Positive and Negative Regulation of the Virulence-Associated Coronafacoyl Phytotoxin in the Potato Common Scab Pathogen Streptomyces scabies.Mol Plant Microbe Interact. 2019 Oct;32(10):1348-1359. doi: 10.1094/MPMI-03-19-0070-R. Epub 2019 Aug 19. Mol Plant Microbe Interact. 2019. PMID: 31107631
-
Characterization of the Coronatine-Like Phytotoxins Produced by the Common Scab Pathogen Streptomyces scabies.Mol Plant Microbe Interact. 2015 Apr;28(4):443-54. doi: 10.1094/MPMI-09-14-0255-R. Mol Plant Microbe Interact. 2015. PMID: 25423263
-
The coronafacoyl phytotoxins: structure, biosynthesis, regulation and biological activities.Antonie Van Leeuwenhoek. 2018 May;111(5):649-666. doi: 10.1007/s10482-017-1009-1. Epub 2018 Jan 6. Antonie Van Leeuwenhoek. 2018. PMID: 29307013 Review.
-
Biosynthesis and regulation of coronatine, a non-host-specific phytotoxin produced by Pseudomonas syringae.Subcell Biochem. 1998;29:321-41. doi: 10.1007/978-1-4899-1707-2_10. Subcell Biochem. 1998. PMID: 9594652 Review.
Cited by
-
Genomic and Metabolomic Analysis of the Potato Common Scab Pathogen Streptomyces scabiei.ACS Omega. 2021 Apr 20;6(17):11474-11487. doi: 10.1021/acsomega.1c00526. eCollection 2021 May 4. ACS Omega. 2021. PMID: 34056303 Free PMC article.
-
Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds.Int J Mol Sci. 2018 Aug 27;19(9):2539. doi: 10.3390/ijms19092539. Int J Mol Sci. 2018. PMID: 30150593 Free PMC article. Review.
-
Critical analysis of polycyclic tetramate macrolactam biosynthetic gene cluster phylogeny and functional diversity.Appl Environ Microbiol. 2024 Jun 18;90(6):e0060024. doi: 10.1128/aem.00600-24. Epub 2024 May 21. Appl Environ Microbiol. 2024. PMID: 38771054 Free PMC article.
-
Beyond Soil-Dwelling Actinobacteria: Fantastic Antibiotics and Where to Find Them.Antibiotics (Basel). 2022 Feb 2;11(2):195. doi: 10.3390/antibiotics11020195. Antibiotics (Basel). 2022. PMID: 35203798 Free PMC article. Review.
-
The virulome of Streptomyces scabiei in response to cello-oligosaccharide elicitors.Microb Genom. 2022 Jan;8(1):000760. doi: 10.1099/mgen.0.000760. Microb Genom. 2022. PMID: 35040428 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

