Genome-wide association study identifies inversion in the CTRB1-CTRB2 locus to modify risk for alcoholic and non-alcoholic chronic pancreatitis
- PMID: 28754779
- PMCID: PMC6145291
- DOI: 10.1136/gutjnl-2017-314454
Genome-wide association study identifies inversion in the CTRB1-CTRB2 locus to modify risk for alcoholic and non-alcoholic chronic pancreatitis
Abstract
Objective: Alcohol-related pancreatitis is associated with a disproportionately large number of hospitalisations among GI disorders. Despite its clinical importance, genetic susceptibility to alcoholic chronic pancreatitis (CP) is poorly characterised. To identify risk genes for alcoholic CP and to evaluate their relevance in non-alcoholic CP, we performed a genome-wide association study and functional characterisation of a new pancreatitis locus.
Design: 1959 European alcoholic CP patients and population-based controls from the KORA, LIFE and INCIPE studies (n=4708) as well as chronic alcoholics from the GESGA consortium (n=1332) were screened with Illumina technology. For replication, three European cohorts comprising 1650 patients with non-alcoholic CP and 6695 controls originating from the same countries were used.
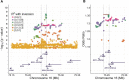
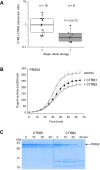
Results: We replicated previously reported risk loci CLDN2-MORC4, CTRC, PRSS1-PRSS2 and SPINK1 in alcoholic CP patients. We identified CTRB1-CTRB2 (chymotrypsin B1 and B2) as a new risk locus with lead single-nucleotide polymorphism (SNP) rs8055167 (OR 1.35, 95% CI 1.23 to 1.6). We found that a 16.6 kb inversion in the CTRB1-CTRB2 locus was in linkage disequilibrium with the CP-associated SNPs and was best tagged by rs8048956. The association was replicated in three independent European non-alcoholic CP cohorts of 1650 patients and 6695 controls (OR 1.62, 95% CI 1.42 to 1.86). The inversion changes the expression ratio of the CTRB1 and CTRB2 isoforms and thereby affects protective trypsinogen degradation and ultimately pancreatitis risk.
Conclusion: An inversion in the CTRB1-CTRB2 locus modifies risk for alcoholic and non-alcoholic CP indicating that common pathomechanisms are involved in these inflammatory disorders.
Keywords: Genome wide association study; chronic pancreatitis; genetic rearrangement.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures


Comment in
-
The CTRB1-CTRB2 risk allele for chronic pancreatitis discovered in European populations does not contribute to disease risk variation in the Chinese population due to near allele fixation.Gut. 2018 Jul;67(7):1368-1369. doi: 10.1136/gutjnl-2017-315180. Epub 2017 Sep 26. Gut. 2018. PMID: 28951524 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
